Bioacoustic research relies on quantifying the structure of acoustic` signals and comparing that structure across behavioral/ecological contexts, groups or species. However, measuring signal structure in a way that fully accounts for the variation in the signals could be a tricky task. Some of the differences that are apparent by visual inspection of spectrograms might not be picked up by some analyses. Hence, choosing the most appropriate analytical approach is a critical step.
The warbleR function
compare_methods() attempts to facilitate method selection. This function
produces graphs (as image files in the working directory) with
spectrograms from 4 signals that allow visual inspection of the
performance of acoustic analysis methods at comparing those signals. The
signals are randomly picked up from the provided data frame (X
argument). The function compares 2 methods at a time. The methods
available are: cross-correlation (using the
warbleR function cross_correlation()),
dynamic time warping on dominant frequency time series (using dfDTW()),
dynamic time warping on fundamental frequency time series (using
ffDTW()), and spectral features (using spectro_analysis()). The graphs also
contain 2 scatterplots (1 for each method) of the acoustic space of all
signals in the input data frame ‘X’. The position of the 4 signals in
the spectrograms is highlighted in the acoustic space scatterplot. In
this way users can directly assess if the distances between signals in
the acoustic space accurately represents the spectrographic similarity
(e.g. how similar their acoustic structure looks in the spectrograms).
This is a short example on how to use the function using the data files included in the package. You need warbleR version 1.1.5 or higher to be able to run the code.
First load the package and save the example sound files in the working directory
library(warbleR)
data(list = c("Phae.long1", "Phae.long2", "Phae.long3", "Phae.long4", "selec.table"))
writeWave(Phae.long1,"Phae.long1.wav")
writeWave(Phae.long2,"Phae.long2.wav")
writeWave(Phae.long3,"Phae.long3.wav")
writeWave(Phae.long4,"Phae.long4.wav")
Then just run the function selecting the 2 methods that you want to
compare. The code below compares cross-correlation (XCORR) and dynamic
time warping on dominant frequency contours (dfDTW). The compared
selections are randomly picked up from the pool of selections in the
input data frame. The argument ‘n’ defines the number of comparisons
(i.e. graphs) to be carried out.
compare_methods(X = selec.table, flim = c(0, 10), bp = c(0, 10),
wl = 300, n = 10, methods = c("XCORR", "dfDTW"))
It should produce 10 image files (in the working directory) that look like these ones:
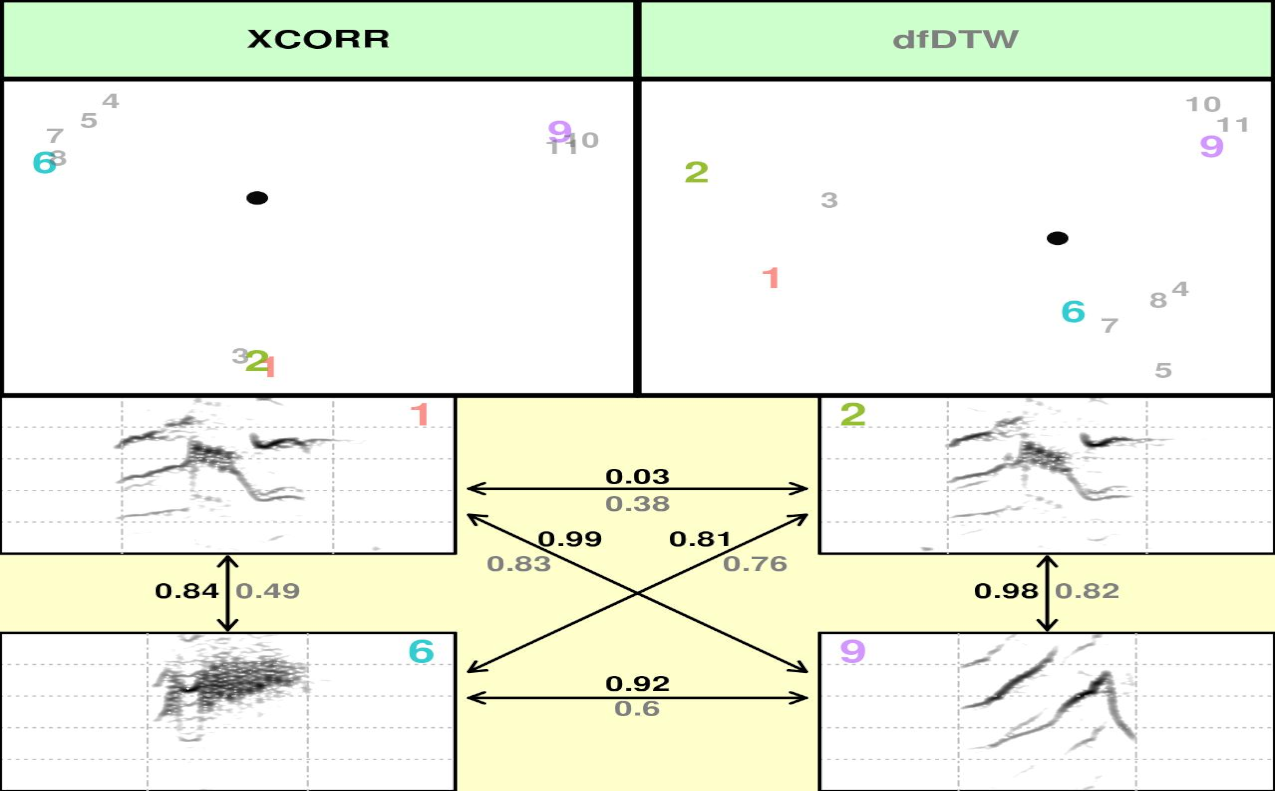
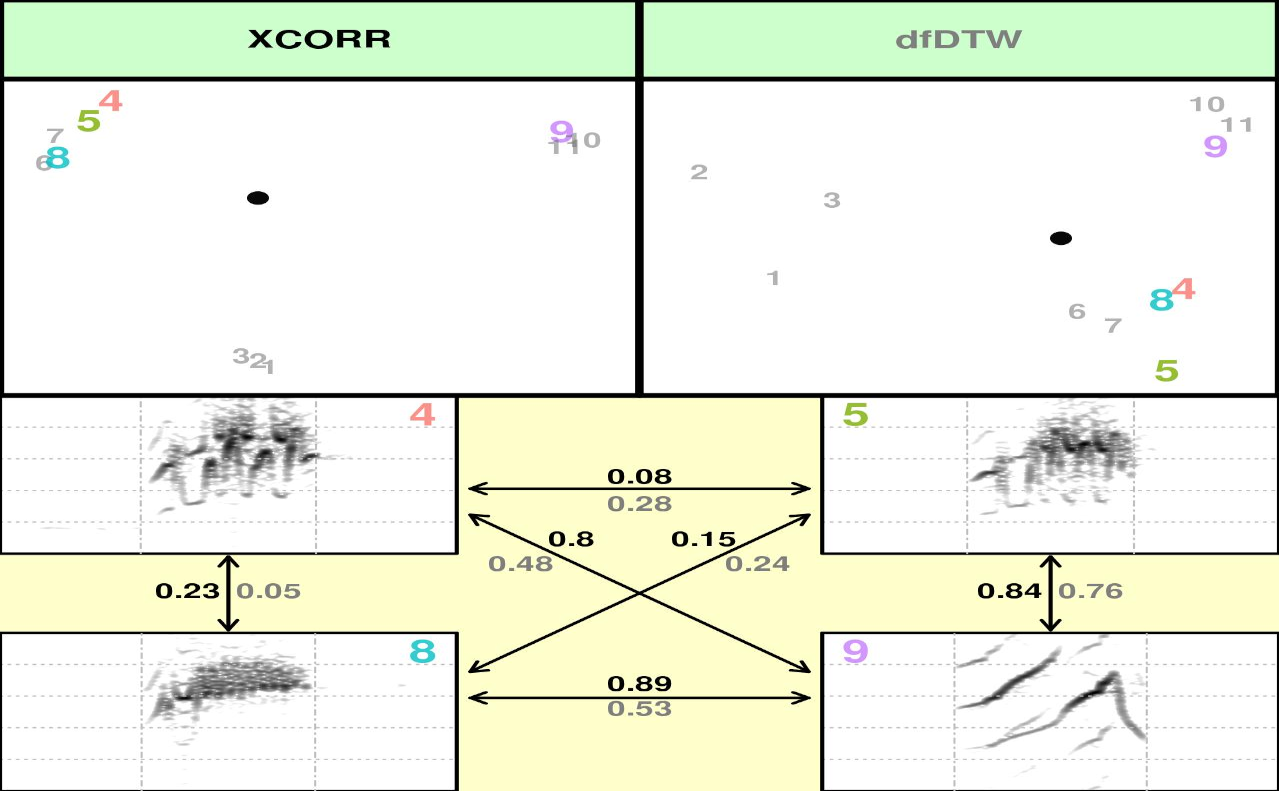
(You can tell where you working directory is found by running getwd())
The acoustic pairwise distance between signals is shown next to the
arrows linking them. The font color of a distance value corresponds to
the font color of the method that generated it, as shown in the
scatterplots (in this case black font represents XCORR distances).
Distances are standardize, being 0 the distance of a signal to itself
and 1 the farthest pairwise distance in the pool of signals. Principal
Component Analysis (princomp() function) is applied to calculate
distances when using spectral features (SP). In that case the first 2
PC’s are used. Classical Multidimensional Scaling (also known as
Principal Coordinates Analysis, cmdscale() function) is used for all
other methods. The file name contains the methods being compared and the
row number of the selections. This function uses internally a modified
version of the spectro() function from
[seewave]((https://cran.r-project.org/package=seewave) package to
create spectrograms. Note that the spectrograms are all plotted with the
same frequency and time scales.
To include the SP method (spectral features) using this data we need
to create a larger data set as the PCA that summarizes the spectral
features needs more units (rows) that variables (columns). So just
create a new selection table repeating 3 times selec.table. It’s a silly
example but it’s just to show how it works, you don’t need to do this if
you have more signals than the number of spectral features in spectro_analysis()
(which at this time is 25)
st2 <- rbind(selec.table, selec.table, selec.table)
#note that the selection labels should be also changed
st2$selec <- 1:nrow(st2)
#now we can compare SP method against XCORR
compare_methods(X = st2, flim = c(0, 10), bp = c(0, 10),
wl = 300, n = 10, methods = c("XCORR", "SP"))
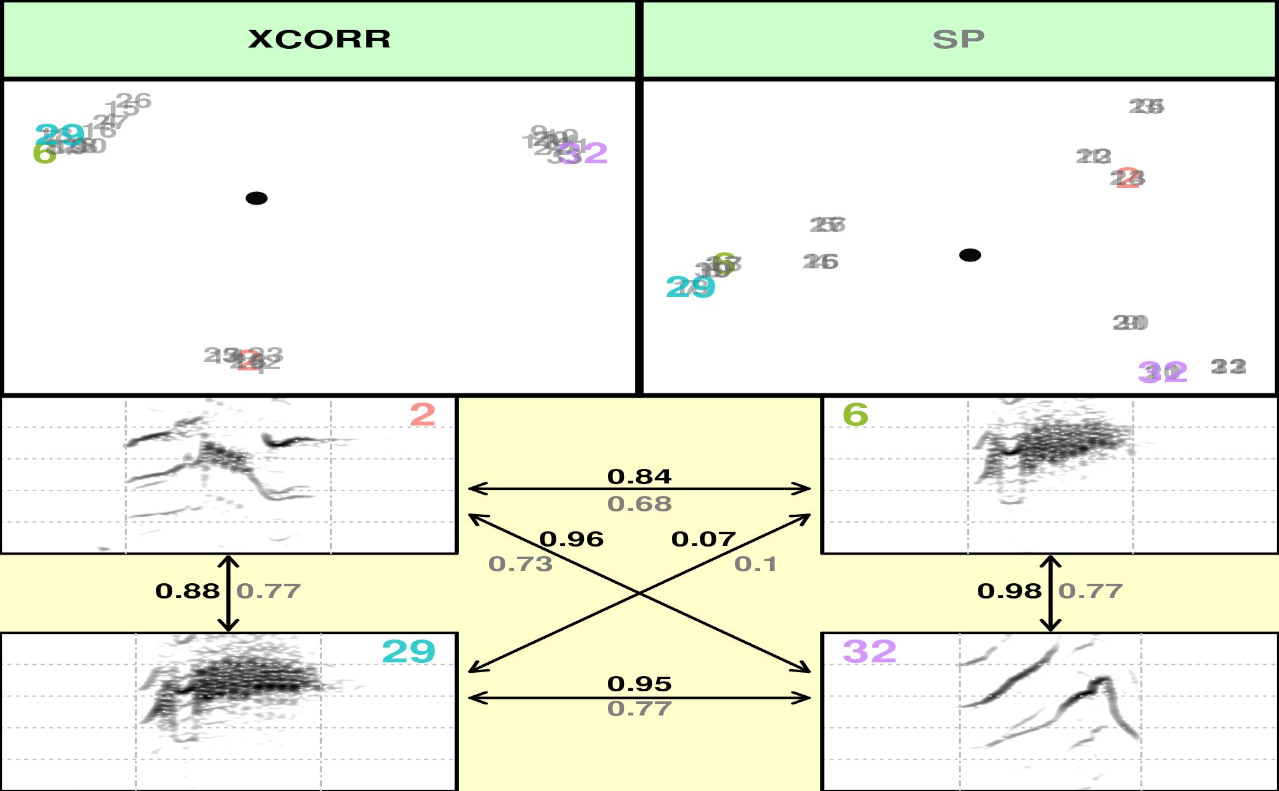
Alternatively, you can provide you own data. This could be helpful for
removing undesired features or to input features obtained elsewhere
(e.g from Raven). To do so, input your data with the sp argument. “SP”
must be included as a method in the ‘methods’ argument. The example
below 1) calculates the spectral features with the spectro_analysis() function,
2) selects only the first 7 columns of the output, and 3) inputs this
data into compare_methods.
sp <- spectro_analysis(selec.table, bp = c(0, 10))
sp <- sp[, 1:7]
compare_methods(X = selec.table, flim = c(0, 10), sp = sp, bp = c(0, 10),
wl = 300, n = 10, methods = c("dfDTW", "SP"))
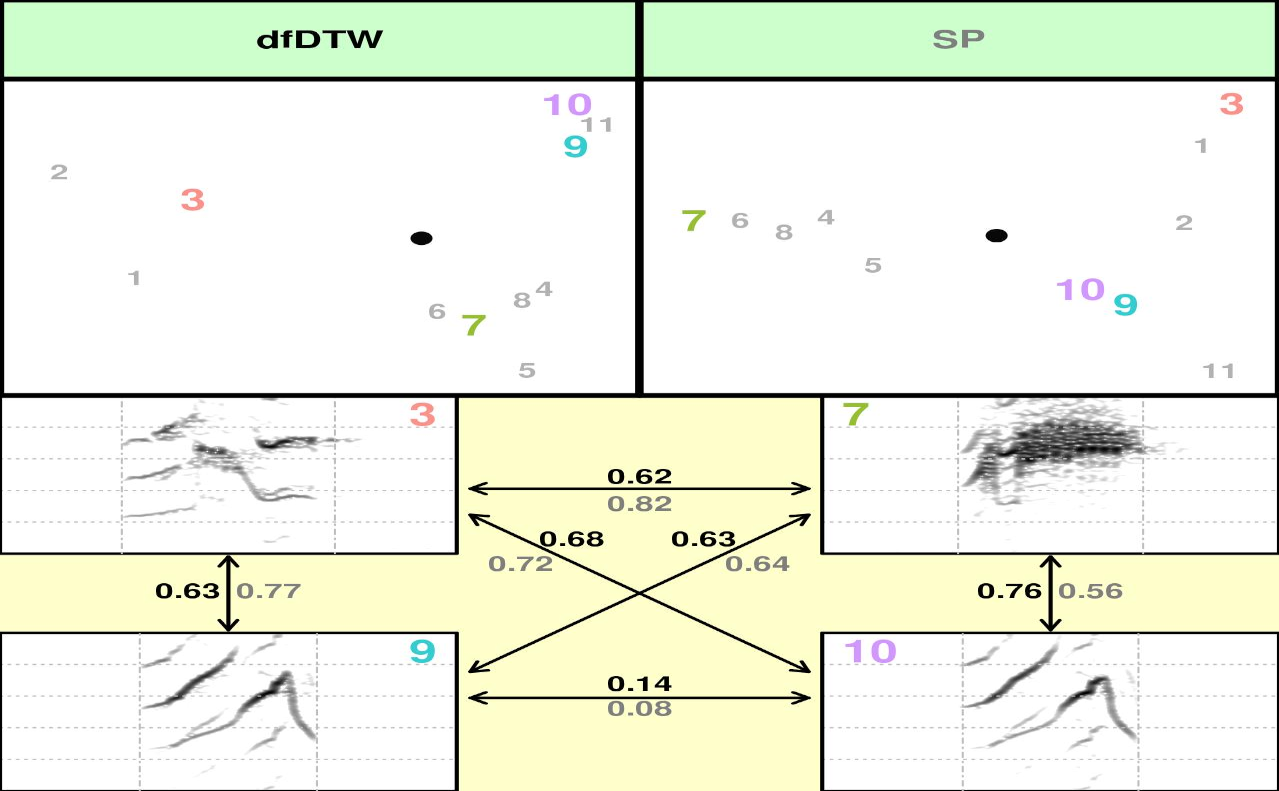
The function has many other arguments to spectrogram parameters (e.g. bandpass, overlap) and visualization settings (e.g. margin, grid, frequency limits). Take a look at the function help document.