The R package dynaSpec can now be installed from github. This is a set of tools to generate dynamic spectrogram visualizations in video format. It is still on the making and new visualizations will be available soon. FFMPEG must be installed in order for this package to work.
To install dynaSpec from github you will need the R package devtools:
# From github
devtools::install_github("maRce10/dynaSpec")
#load package
library(dynaSpec)
Installation of external dependencies can be tricky on operating systems other than Linux. An alternative option is to run the package through google colab. This colab notebook explain how to do that step-by-step.
Examples
To run the following examples you will also need to load a few more packages:
#load package
library(warbleR)
library(viridis)
library(tuneR)
library(seewave)
The function scrolling_spectro() (the only function so far) creates
‘.mp4’ video files of the spectrogram of an input ‘wave’ object that
moves from right to left. For instance, this is a dynamic spectrogram of
a canyon wren song with a viridis color palette:
data("canyon_wren")
scrolling_spectro(wave = canyon_wren, wl = 300,
t.display = 1.7, pal = viridis,
grid = FALSE, flim = c(1, 9),
width = 1000, height = 500,
res = 120, file.name = "default.mp4")
This is the same clip but with a black and white spectrogram:
scrolling_spectro(wave = canyon_wren, wl = 300,
t.display = 1.7, pal = reverse.gray.colors.1,
grid = FALSE, flim = c(1, 9),
width = 1000, height = 500,
res = 120, file.name = "black_and_white.mp4",
collevels = seq(-100, 0, 5))
A spectrogram with black background (colbg = “black”):
scrolling_spectro(wave = canyon_wren, wl = 300,
t.display = 1.7, pal = viridis,
grid = FALSE, flim = c(1, 9),
width = 1000, height = 500, res = 120,
file.name = "black.mp4", colbg = "black")
Slow down to 1/2 speed (speed = 0.5) with a oscillogram at the bottom (osc = TRUE):
scrolling_spectro(wave = canyon_wren, wl = 300,
t.display = 1.7, pal = viridis,
grid = FALSE, flim = c(1, 9),
width = 1000, height = 500, res = 120,
file.name = "slow.mp4", colbg = "black",
speed = 0.5, osc = TRUE,
colwave = "#31688E99")
Long-billed hermit song at 1/5 speed (speed = 0.5), removing axes and looping 3 times (loop = 3:
data("Phae.long4")
scrolling_spectro(wave = Phae.long4, wl = 300,
t.display = 1.7, ovlp = 90, pal = magma,
grid = FALSE, flim = c(1, 10), width = 1000,
height = 500, res = 120, collevels = seq(-50, 0, 5),
file.name = "no_axis.mp4", colbg = "black",
speed = 0.2, axis.type = "none", loop = 3)
Visualizing a northern nightingale wren recording from xeno-canto using a custom color palette:
ngh_wren <- read_wave("https://www.xeno-canto.org/518334/download")
custom_pal <- colorRampPalette( c("#2d2d86", "#2d2d86", reverse.terrain.colors(10)[5:10]))
scrolling_spectro(wave = ngh_wren, wl = 600,
t.display = 3, ovlp = 95, pal = custom_pal,
grid = FALSE, flim = c(2, 8), width = 700,
height = 250, res = 100, collevels = seq(-40, 0, 5),
file.name = "../nightingale_wren.mp4", colbg = "#2d2d86", lcol = "#FFFFFFE6")
Spix’s disc-winged bat inquiry call slow down (speed = 0.05):
data("thyroptera.est")
# extract one call
thy_wav <- attributes(thyroptera.est)$wave.objects[[12]]
# add silence at both "sides""
thy_wav <- pastew(tuneR::silence(duration = 0.05,
samp.rate = thy_wav@samp.rate, xunit = "time"),
thy_wav, output = "Wave")
thy_wav <- pastew(thy_wav, tuneR::silence(duration = 0.04,
samp.rate = thy_wav@samp.rate, xunit = "time"),
output = "Wave")
scrolling_spectro(wave = thy_wav, wl = 400,
t.display = 0.08, ovlp = 95, pal = inferno,
grid = FALSE, flim = c(12, 37), width = 700,
height = 250, res = 100, collevels = seq(-40, 0, 5),
file.name = "thyroptera_osc.mp4", colbg = "black", lcol = "#FFFFFFE6",
speed = 0.05, fps = 200, buffer = 0, loop = 4, lty = 1,
osc = TRUE, colwave = inferno(10, alpha = 0.9)[3])
Further customization
The argument ‘spectro.call’ allows to insert customized spectrogram
visualizations. For instance, the following code makes use of the
color_spectro() function from
warbleR to highlight
vocalizations from male and female house wrens with different colors
(after downloading the selection table and sound file from figshare):
# get house wren male female duet recording
hs_wren <- read_wave("https://ndownloader.figshare.com/files/22722101")
# and extended selection table
st <- read.csv("https://ndownloader.figshare.com/files/22722404")
# create color column
st$colors <- c("green", "yellow")
# highlight selections
color.spectro(wave = hs_wren, wl = 200, ovlp = 95, flim = c(1, 13),
collevels = seq(-55, 0, 5), dB = "B", X = st, col.clm = "colors",
base.col = "black", t.mar = 0.07, f.mar = 0.1, strength = 3,
interactive = NULL, bg.col = "black")
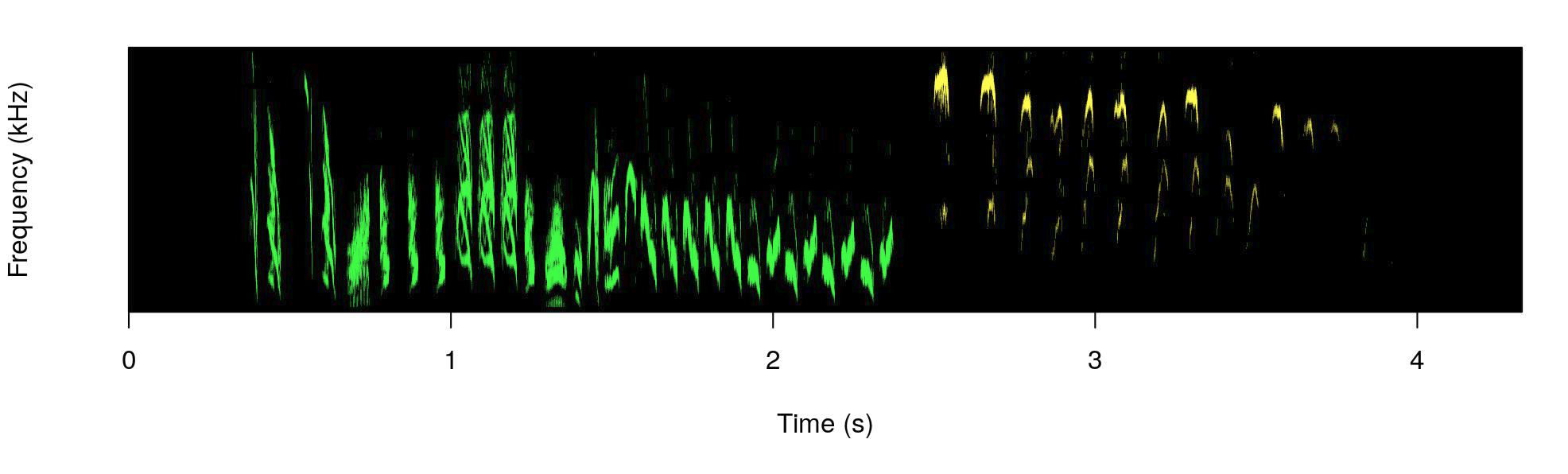
The male part is shown in green and the female part in yellow.
We can wrap the color_spectro() call using the call() function form
base R and input that into scrolling_spectro() using the argument
‘spectro.call’:
# save call
sp_cl <- call("color.spectro", wave = hs_wren, wl = 200, ovlp = 95,
flim = c(1, 13), collevels = seq(-55, 0, 5), strength = 3,
dB = "B", X = st, col.clm = "colors", base.col = "black",
t.mar = 0.07, f.mar = 0.1, interactive = NULL, bg.col = "black")
# create dynamic spectrogram
scrolling_spectro(wave = hs_wren, wl = 512,
t.display = 1.2, pal = reverse.gray.colors.1,
grid = FALSE, flim = c(1, 13), loop = 3,
width = 1000, height = 500, res = 120,
collevels = seq(-100, 0, 1), spectro.call = sp_cl, fps = 60,
file.name = "yellow_and_green.mp4")
This option can be mixed with any of the other customizations in the function, as adding an oscillogram:
# create dynamic spectrogram
scrolling_spectro(wave = hs_wren, wl = 512, osc = TRUE,
t.display = 1.2, pal = reverse.gray.colors.1,
grid = FALSE, flim = c(1, 13), loop = 3,
width = 1000, height = 500, res = 120,
collevels = seq(-100, 0, 1),
spectro.call = sp_cl, fps = 60,
file.name = "yellow_and_green_oscillo.mp4")
A viridis color palette:
st$colors <- viridis(10)[c(3, 8)]
sp_cl <- call("color.spectro", wave = hs_wren, wl = 200,
ovlp = 95, flim = c(1, 13), collevels = seq(-55, 0, 5),
dB = "B", X = st, col.clm = "colors",
base.col = "white", t.mar = 0.07, f.mar = 0.1,
strength = 3, interactive = NULL)
# create dynamic spectrogram
scrolling_spectro(wave = hs_wren, wl = 200, osc = TRUE,
t.display = 1.2, pal = reverse.gray.colors.1,
grid = FALSE, flim = c(1, 13), loop = 3,
width = 1000, height = 500, res = 120,
collevels = seq(-100, 0, 1), colwave = viridis(10)[c(9)],
spectro.call = sp_cl, fps = 60,
file.name = "viridis.mp4")
Or simply a gray scale:
st$colors <- c("gray", "gray49")
sp_cl <- call("color.spectro", wave = hs_wren, wl = 200, ovlp = 95, flim = c(1, 13),
collevels = seq(-55, 0, 5), dB = "B", X = st, col.clm = "colors",
base.col = "white", t.mar = 0.07, f.mar = 0.1, strength = 3,
interactive = NULL)
# create dynamic spectrogram
scrolling_spectro(wave = hs_wren, wl = 512, osc = TRUE,
t.display = 1.2, pal = reverse.gray.colors.1,
grid = FALSE, flim = c(1, 13), loop = 3,
width = 1000, height = 500, res = 120,
collevels = seq(-100, 0, 1),
spectro.call = sp_cl, fps = 60,
file.name = "gray.mp4")
The ‘spectro.call’ argument can also be used to add annotations. To do
this we need to wrap up both the spectrogram function and the annotation
functions (i.e. text(), lines()) in a single function and then save
the call to that function:
# create color column
st$colors <- viridis(10)[c(3, 8)]
# create label column
st$labels <- c("male", "female")
# shrink end of second selection (purely aesthetics)
st$end[2] <- 3.87
# function to highlight selections
ann_fun <- function(wave, X){
# print spectrogram
color.spectro(wave = wave, wl = 200,
ovlp = 95, flim = c(1, 18.6), collevels = seq(-55, 0, 5),
dB = "B", X = X, col.clm = "colors",
base.col = "white", t.mar = 0.07, f.mar = 0.1,
strength = 3, interactive = NULL)
# annotate each selection in X
for(e in 1:nrow(X)){
# label
text(x = X$start[e] + ((X$end[e] - X$start[e]) / 2),
y = 16.5, labels = X$labels[e], cex = 3.3,
col = adjustcolor(X$colors[e], 0.6))
# line
lines(x = c(X$start[e], X$end[e]), y = c(14.5, 14.5),
lwd = 6, col = adjustcolor("gray50", 0.3))
}
}
# save call
ann_cl <- call("ann_fun", wave = hs_wren, X = st)
# create annotated dynamic spectrogram
scrolling_spectro(wave = hs_wren, wl = 200, t.display = 1.2,
grid = FALSE, flim = c(1, 18.6), loop = 3,
width = 1000, height = 500, res = 200,
collevels = seq(-100, 0, 1), speed = 0.5,
spectro.call = ann_cl, fps = 120,
file.name = "../viridis_annotated.mp4")
Finally, the argument ‘annotation.call’ can be used to add static labels
(i.e. non-scrolling). It works similar to ‘spectro.call’, but requires a
call from text(). This let users customize things as size, color,
position, font, and additional arguments taken by text(). The call
should also include the argmuents ‘start’ and ‘end’ to indicate the time
at which the labels are displayed (in s). ‘fading’ is optional and
allows fade-in and fade-out effects on labels (in s as well). The
following code downloads a recording containing several frog species
recorded in Costa Rica from figshare, cuts a clip including two species
and labels it with a single label:
# read data from figshare
frogs <- read_wave("https://ndownloader.figshare.com/files/22829075")
# cut a couple of species
shrt_frgs <- cutw(frogs, from = 35.3, to = 50.5, output = "Wave")
# make annotation call
ann_cll <- call("text", x = 0.25, y = 0.87,
labels = "Frog calls", cex = 1, start = 0.2, end = 14,
col = "#FFEA46CC", font = 3, fading = 0.6)
# create dynamic spectro
scrolling_spectro(wave = shrt_frgs, wl = 512, ovlp = 95,
t.display = 1.1, pal = cividis,
grid = FALSE, flim = c(0, 5.5), loop = 3,
width = 1200, height = 550, res = 200,
collevels = seq(-40, 0, 5), lcol = "#FFFFFFCC",
colbg = "black", fps = 60, file.name = "../frogs.mp4",
osc = TRUE, height.prop = c(3, 1), colwave = "#31688E",
lty = 3, annotation.call = ann_cll)
The argument accepts more than one labels as in a regular text() call.
In that case ‘start’ and ‘end’ values should be supplied for each label:
# make annotation call for 2 annotations
ann_cll <- call("text", x = 0.25, y = 0.87,
labels = c("Dendropsophus ebraccatus", "Eleutherodactylus coqui"),
cex = 1, start = c(0.4, 7),
end = c(5.5, 14.8), col = "#FFEA46CC", font = 3, fading = 0.6)
# create dynamic spectro
scrolling_spectro(wave = shrt_frgs, wl = 512, ovlp = 95,
t.display = 1.1, pal = cividis,
grid = FALSE, flim = c(0, 5.5), loop = 3,
width = 1200, height = 550, res = 200,
collevels = seq(-40, 0, 5), lcol = "#FFFFFFCC", colbg = "black",
fps = 60, file.name = "../frogs_sp_labels.mp4", osc = TRUE,
height.prop = c(3, 1),colwave = "#31688E", lty = 3,
annotation.call = ann_cll)
Please cite dynaSpec as follows:
Araya-Salas M. (2020), dynaSpec: dynamic spectrogram visualizations in R. R package version 1.0.0.
Session information
## R version 4.3.2 (2023-10-31)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 22.04.2 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## time zone: America/Costa_Rica
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] warbleR_1.1.30 NatureSounds_1.0.4 knitr_1.45 seewave_2.2.3
## [5] tuneR_1.4.6
##
## loaded via a namespace (and not attached):
## [1] cli_3.6.2 rlang_1.1.3 xfun_0.41 jsonlite_1.8.8
## [5] rjson_0.2.21 RCurl_1.98-1.14 dtw_1.23-1 fftw_1.0-8
## [9] htmltools_0.5.7 brio_1.1.4 rmarkdown_2.25 evaluate_0.23
## [13] MASS_7.3-55 bitops_1.0-7 fastmap_1.1.1 yaml_2.3.8
## [17] compiler_4.3.2 Rcpp_1.0.12 testthat_3.2.1 pbapply_1.7-2
## [21] rstudioapi_0.15.0 digest_0.6.34 R6_2.5.1 signal_1.8-0
## [25] parallel_4.3.2 magrittr_2.0.3 tools_4.3.2 proxy_0.4-27