
package overview
suwo: access nature media repositories
Jorge Elizondo, Marcelo Araya-Salas & Alejandro Rico-Guevara
2026-02-05
Source:vignettes/suwo.Rmd
suwo.Rmd
The suwo package aims
to simplify the retrieval of nature media (mostly photos, audio files
and videos) across multiple online biodiversity databases. This vignette
provides an overview of the package’s core querying functions, the
searching and downloading of media files, and the compilation of
metadata from various sources. For detailed information on each
function, please refer to the function
reference or use the help files within R (e.g.,
?query_gbif).
Intended use and responsible practices
This package is designed exclusively for non-commercial, scientific purposes, including research, education, and conservation. Any commercial use of the data or media retrieved through this package is strictly prohibited unless explicit, separate permission is granted directly from the original source platforms and rights holders. Users are obligated to comply with the specific terms of service and data use policies of each source database, which often mandate attribution and similarly restrict commercial application. The package developers assume no liability for misuse of the retrieved data or violations of third-party terms of service.
Basic workflow for obtaining nature media files
Obtaining nature media using suwo follows a basic sequence. The following diagram illustrates this workflow and the main functions involved:
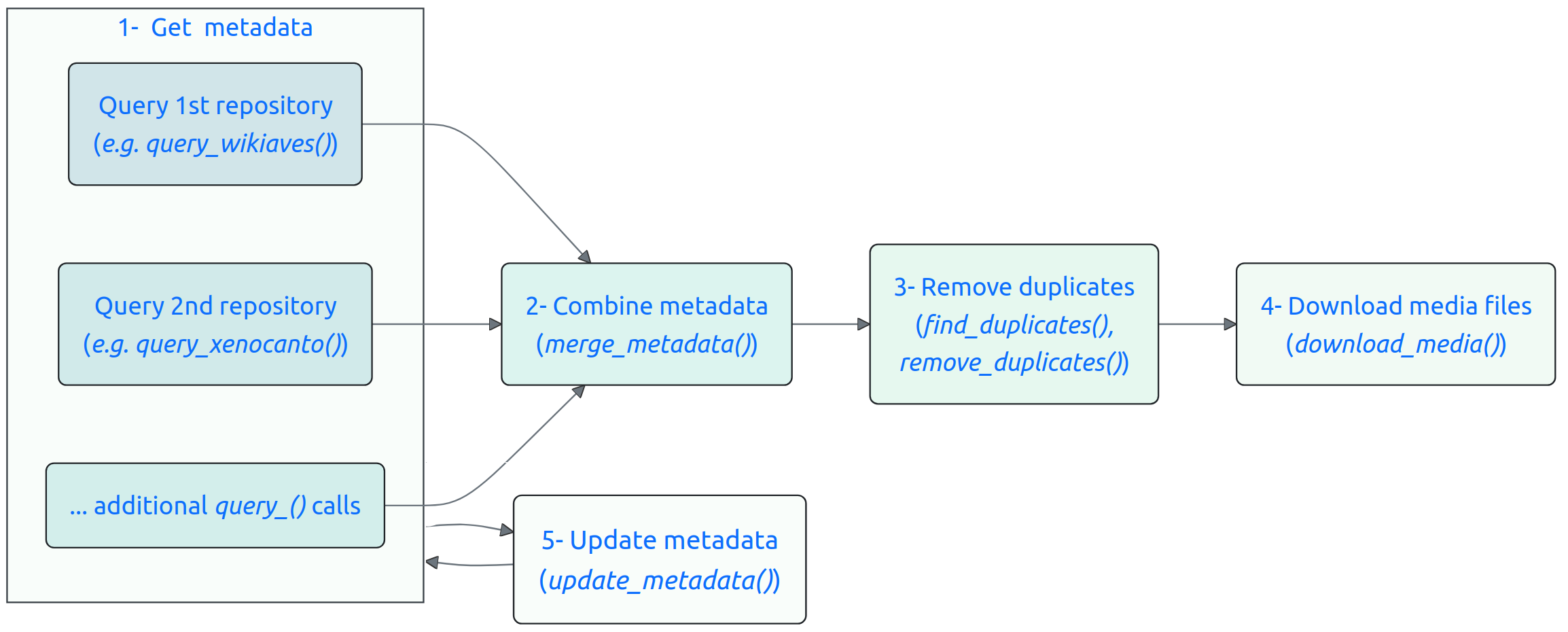
Here is a description of each step:
Obtain metadata:
- Queries regarding a species are submitted through one of the
available query functions (
query_repo_name()) that connect to five different online repositories (Xeno-Canto, Inaturalist, GBIF, Macaulay Library and WikiAves). The output of these queries is a data frame containing metadata associated with the media files (e.g., species name, date, location, etc, see below).
Curate metadata:
If multiple repositories are queried, the resulting metadata data frames can be merged into a single data frame using the merge_metadata() function.
Check for duplicate records in their datasets using the find_duplicates() function. Candidate duplicated entries are identified based on matching species name, country, date, user name, and geographic coordinates. User can double check the candidate duplicates and decide which records to keep, which can be done with remove_duplicates().
Download the media files associated with the metadata using the download_media() function.
Users can update their datasets with new records using the update_metadata() function.
Obtaining metadata: the query functions
The following table summarizes the available suwo query functions and the types of metadata they retrieve:
| Function | Repository | URL link | File types | Requires api key | Taxonomic level | Geographic coverage | Taxonomic coverage | Other features |
|---|---|---|---|---|---|---|---|---|
| query_gbif | GBIF | https://www.gbif.org/ | image, sound, video, interactive resource | No | Species | Global | All life | Specify query by data base |
| query_inaturalist | iNaturalist | https://www.inaturalist.org/ | image, sound | No | Species | Global | All life | |
| query_macaulay | Macaulay Library | https://www.macaulaylibrary.org/ | image, sound, video | No | Species | Global | Mostly birds but also other vertebrates and invertebrates | Interactive |
| query_wikiaves | WikiAves | https://www.wikiaves.com.br/ | image, sound | No | Species | Brazil | Birds | |
| query_xenocanto | Xeno-Canto | https://www.xeno-canto.org/ | sound | Yes | Species, subspecies, genus, family, group | Global | Birds, frogs, non-marine mammals and grasshoppers | Specify query by taxonomy, geographic range and dates |
These are some example queries:
- Images of Sarapiqui Heliconia (Heliconia sarapiquensis) from iNaturalist (we print the first 4 rows of each output data frame):
# Load suwo package
library(suwo)
h_sarapiquensis <- query_inaturalist(species = "Heliconia sarapiquensis",
format = "image")
[32m✔
[39m Obtaining metadata (29 matching records found) 🥇:
head(h_sarapiquensis, 4)| repository | format | key | species | date | time | user_name | country | locality | latitude | longitude | file_url | file_extension |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| iNaturalist | image | 330280680 | Heliconia sarapiquensis | 2025-12-08 | 13:47 | Carlos g Velazco-Macias | NA | 10.159645,-83.9378766667 | 10.15964 | -83.93788 | https://inaturalist-open-data.s3.amazonaws.com/photos/598874322/original.jpg | jpeg |
| iNaturalist | image | 330280680 | Heliconia sarapiquensis | 2025-12-08 | 13:47 | Carlos g Velazco-Macias | NA | 10.159645,-83.9378766667 | 10.15964 | -83.93788 | https://inaturalist-open-data.s3.amazonaws.com/photos/598874346/original.jpg | jpeg |
| iNaturalist | image | 330280680 | Heliconia sarapiquensis | 2025-12-08 | 13:47 | Carlos g Velazco-Macias | NA | 10.159645,-83.9378766667 | 10.15964 | -83.93788 | https://inaturalist-open-data.s3.amazonaws.com/photos/598874381/original.jpg | jpeg |
| iNaturalist | image | 263417773 | Heliconia sarapiquensis | 2025-02-28 | 14:23 | Original Madness | NA | 10.163116739,-83.9389050007 | 10.16312 | -83.93891 | https://inaturalist-open-data.s3.amazonaws.com/photos/473219810/original.jpeg | jpeg |
- Harpy eagles (Harpia harpyja) audio recordings from WikiAves:
h_harpyja <- query_wikiaves(species = "Harpia harpyja", format = "sound")
head(h_harpyja, 4)| repository | format | key | species | date | time | user_name | country | locality | latitude | longitude | file_url | file_extension |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wikiaves | sound | 25867 | Harpia harpyja | NA | NA | Gustavo Pedersoli | Brazil | Alta Floresta/MT | NA | NA | https://s3.amazonaws.com/media.wikiaves.com.br/recordings/52/25867_a73f0e8da2179e82af223ff27f74a912.mp3 | mp3 |
| Wikiaves | sound | 2701424 | Harpia harpyja | 2020-10-20 | NA | Bruno Lima | Brazil | Itanhaém/SP | NA | NA | https://s3.amazonaws.com/media.wikiaves.com.br/recordings/1072/2701424_e0d533b952b64d6297c4aff21362474b.mp3 | mp3 |
| Wikiaves | sound | 878999 | Harpia harpyja | 2013-03-20 | NA | Thiago Silveira | Brazil | Alta Floresta/MT | NA | NA | https://s3.amazonaws.com/media.wikiaves.com.br/recordings/878/878999_c1f8f4ba81fd597548752e92f1cdba50.mp3 | mp3 |
| Wikiaves | sound | 3027120 | Harpia harpyja | 2016-06-20 | NA | Ciro Albano | Brazil | Camacan/BA | NA | NA | https://s3.amazonaws.com/media.wikiaves.com.br/recordings/7203/3027120_5148ce0fed5fe99aba7c65b2f045686a.mp3 | mp3 |
- Common raccoon (Procyon lotor) videos from GBIF:
p_lotor <- query_gbif(species = "Procyon lotor", format = "video")
head(p_lotor, 4)| repository | format | key | species | date | time | user_name | country | locality | latitude | longitude | file_url | file_extension |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GBIF | video | 3501153129 | Procyon lotor | 2015-07-21 | NA | NA | Luxembourg | NA | 49.7733 | 5.94092 | https://archimg.mnhn.lu/Observations/Taxons/Biomonitoring/063_094_S2_K2_20150721_063004AM.mp4 | m4a |
| GBIF | video | 3501153135 | Procyon lotor | 2015-07-04 | NA | NA | Luxembourg | NA | 49.7733 | 5.94092 | https://archimg.mnhn.lu/Observations/Taxons/Biomonitoring/063_094_S2_K1_20150704_072418AM.mp4 | m4a |
| GBIF | video | 3501153159 | Procyon lotor | 2015-07-04 | NA | NA | Luxembourg | NA | 49.7733 | 5.94092 | https://archimg.mnhn.lu/Observations/Taxons/Biomonitoring/063_094_S2_K1_20150704_072402AM.mp4 | m4a |
| GBIF | video | 3501153162 | Procyon lotor | 2015-07-04 | NA | NA | Luxembourg | NA | 49.7733 | 5.94092 | https://archimg.mnhn.lu/Observations/Taxons/Biomonitoring/063_094_S2_K1_20150704_072346AM.mp4 | m4a |
By default all query function return the 13 most basic metadata fields associated with the media files. Here is the definition of each field:
- repository: Name of the repository
- format: Type of media file (e.g., sound, photo, video)
- key: Unique identifier of the media file in the repository
- species: Species name associated with the media file (Note taxonomic authority may vary among repositories)
- date*: Date when the media file was recorded/photographed (in YYYY-MM-DD format or YYYY if only year is available)
- time*: Time when the media file was recorded/photographed (in HH:MM format)
- user_name*: Name of the user who uploaded the media file
- country*: Country where the media file was recorded/photographed
- locality*: Locality where the media file was recorded/photographed
- latitude*: Latitude of the location where the media file was recorded/photographed (in decimal degrees)
- longitude*: Longitude of the location where the media file was recorded/photographed (in decimal degrees)
- file_url: URL link to the media file (used to download media files)
- file_extension: Extension of the media file (e.g., .mp3, .jpg, .mp4)
* Can contain missing values (NAs)
Users can also download all available metadata by setting the
argument all_data = TRUE. These are the additional metadata
fields, on top of the basic fields, that are retrieved by each query
function:
| Function | Additional data |
|---|---|
| query_gbif | datasetkey, publishingorgkey, installationkey, hostingorganizationkey, publishingcountry, protocol, lastcrawled, lastparsed, crawlid, basisofrecord, occurrencestatus, taxonkey, kingdom_code, phylum_code, class_code, order_code, family_key, genus_code, species_code, acceptedtaxonkey, scientificnameauthorship, acceptedscientificname, kingdom, phylum, order, family, genus, genericname, specific_epithet, taxonrank, taxonomicstatus, iucnredlistcategory, continent, year, month, day, startdayofyear, enddayofyear, lastinterpreted, license, organismquantity, organismquantitytype, issequenced, isincluster, datasetname, recordist, identifiedby, samplingprotocol, geodeticdatum, class, countrycode, gbifregion, publishedbygbifregion, recordnumber, identifier, habitat, verbatimeventdate, institutionid, dynamicproperties, verbatimcoordinatesystem, eventremarks, collectioncode, gbifid, occurrenceid, institutioncode, identificationqualifier, media_type, page, state_province, comments |
| query_inaturalist | quality_grade, taxon_geoprivacy, uuid, cached_votes_total, identifications_most_agree, species_guess, identifications_most_disagree, positional_accuracy, comments_count, site_id, created_time_zone, license_code, observed_time_zone, public_positional_accuracy, oauth_application_id, created_at, description, time_zone_offset, observed_on, observed_on_string, updated_at, captive, faves_count, num_identification_agreements, identification_disagreements_count, map_scale, uri, community_taxon_id, owners_identification_from_vision, identifications_count, obscured, num_identification_disagreements, geoprivacy, spam, mappable, identifications_some_agree, place_guess, id, license_code_1, attribution, hidden, offset |
| query_macaulay | common_name, background_species, caption, year, month, day, country_state_county, state_province, county, age_sex, behavior, playback, captive, collected, specimen_id, home_archive_catalog_number, recorder, microphone, accessory, partner_institution, ebird_checklist_id, unconfirmed, air_temp_c, water_temp_c, media_notes, observation_details, parent_species, species_code, taxon_category, taxonomic_sort, recordist_2, average_community_rating, number_of_ratings, asset_tags, original_image_height, original_image_width |
| query_wikiaves | user_id, species_code, common_name, repository_id, verified, locality_id, number_of_comments, likes, visualizations, duration |
| query_xenocanto | genus, specific_epithet, subspecies, taxonomic_group, english_name, altitude, vocalization_type, sex, stage, method, url, uploaded_file, license, quality, length, upload_date, other_species, comments, animal_seen, playback_used, temp, regnr, auto, recorder, microphone, sampling_rate, sonogram_small, sonogram_med, sonogram_large, sonogram_full, oscillogram_small, oscillogram_med, oscillogram_large, sonogram |
Obtaining raw data
By default the package standardizes the information in the basic
fields (detailed above) in order to facilitate the compilation of
metadata from multiple repositories. However, in some cases this may
result in loss of information. For instance, some repositories allow
users to provide “morning” as a valid time value, which are converted
into NAs by suwo. In such
cases, users can retrieve the original data by setting the
raw_data = TRUE in the query functions and/or global
options (options(raw_data = TRUE)). Note that subsequent
data manipulation functions (e.g., merge_metadata(),
find_duplicates(),
etc) will not work as the basic fields are not standardized.
The code above examplifies the most common use of query functions, which applies also to the function query_gbif(). The following sections provide more details on the two query functions that require special considerations: query_macaulay() and query_xenocanto().
query_macaulay()
Interactive retrieval of metadata
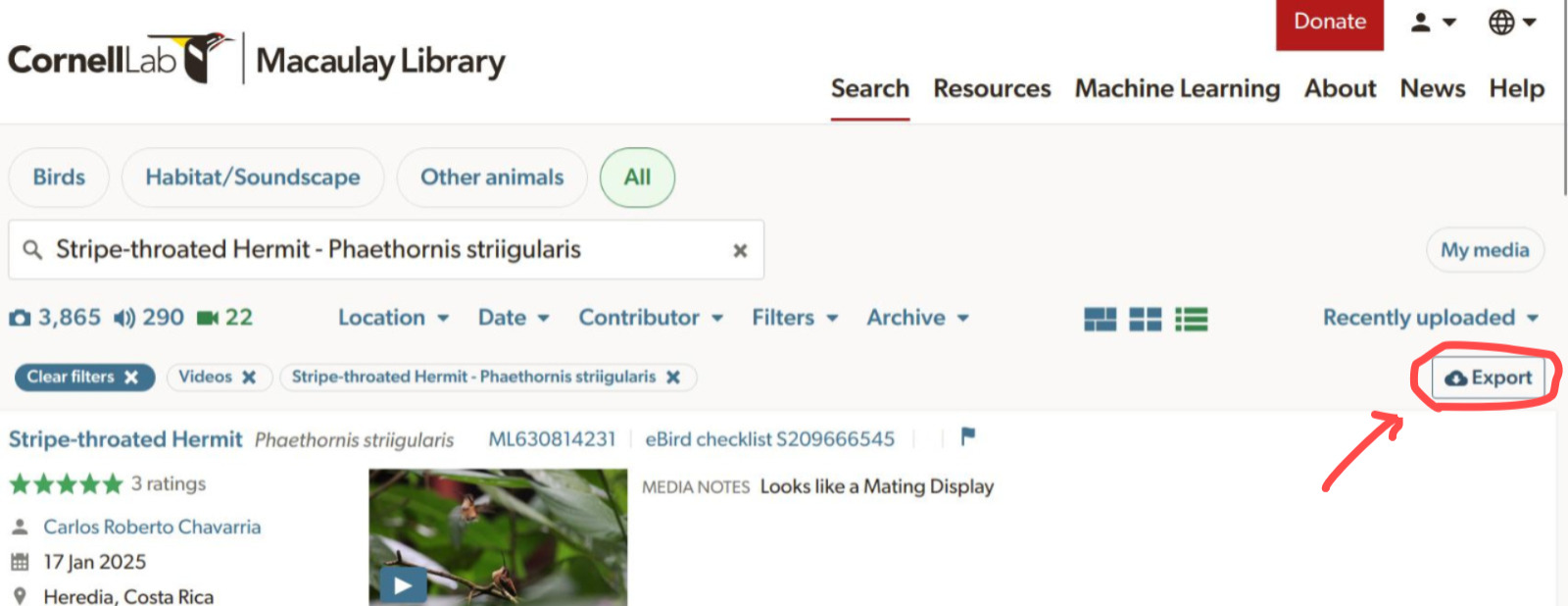
query_macaulay() is the only interactive function. This means that when users run a query the function opens a browser window to the Macaulay Library’s search page, where the users must download a .csv file with the metadata. Here is a example of a query for strip-throated hermit (Phaethornis striigularis) videos:
p_striigularis <- query_macaulay(species = "Phaethornis striigularis",
format = "video")Users must click on the “Export” button to save the .csv file with the metadata:
Note that for bird species the species name must be valid according
to the Macaulay Library taxonomy (which follows the Clements checklist).
For non-bird species users must use the argument
taxon_code. The species taxon code can be found by running
a search at the Macaulay Library’s
search page and checking the URL of the species page. For instance,
the taxon code for jaguar (Panthera onca) is “t-11032765”:
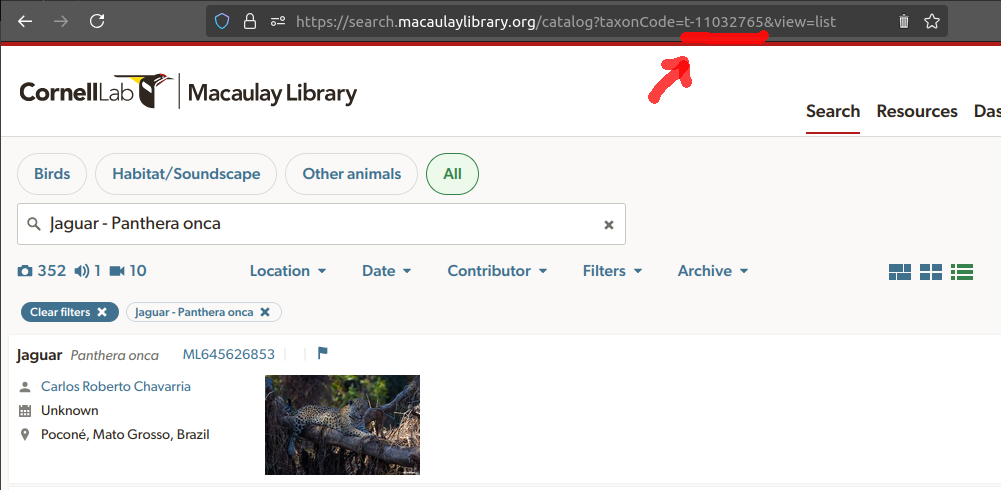
Once you have the taxon code, you can run the query as follows:
jaguar <- query_macaulay(taxon_code = "t-11032765",
format = "video")Here are some tips for using this function properly:
- Valid bird species names can be checked at
suwo:::ml_taxon_code$SCI_NAME - The exported csv file must be saved in the directory specified by
the argument
pathof the function (default is the current working directory) - If the file is saved overwriting a pre-existing file (i.e. same file name) the function will not detect it
- The function will not proceed until the file is saved
- Users must log in to the Macaulay Library/eBird account in order to access large batches of observations
After saving the file, the function will read the file and return a data frame with the metadata. Here we print the first 4 rows of the output data frame:
head(p_striigularis, 4)| repository | format | key | species | date | time | user_name | country | locality | latitude | longitude | file_url | file_extension |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Macaulay Library | video | 630814231 | Phaethornis striigularis | 2025-01-17 | 09:23 | Carlos Roberto Chavarria | Costa Rica | Tirimbina Rainforest Center | 10.41562 | -84.12078 | https://cdn.download.ams.birds.cornell.edu/api/v1/asset/630814231/ | mp4 |
| Macaulay Library | video | 628258211 | Phaethornis striigularis | 2024-12-13 | 14:26 | Russell Campbell | Costa Rica | Reserva El Copal (Tausito) | 9.78404 | -83.75147 | https://cdn.download.ams.birds.cornell.edu/api/v1/asset/628258211/ | mp4 |
| Macaulay Library | video | 628258206 | Phaethornis striigularis | 2024-12-13 | 14:26 | Russell Campbell | Costa Rica | Reserva El Copal (Tausito) | 9.78404 | -83.75147 | https://cdn.download.ams.birds.cornell.edu/api/v1/asset/628258206/ | mp4 |
| Macaulay Library | video | 614437320 | Phaethornis striigularis | 2022-10-15 | 14:40 | Josep Del Hoyo | Costa Rica | Laguna Lagarto Lodge | 10.68515 | -84.18112 | https://cdn.download.ams.birds.cornell.edu/api/v1/asset/614437320/ | mp4 |
Bypassing record limit
Even if logged in, a maximum of 10000 records per query can be
returned. This can be bypassed by using the argument dates
to split the search into a sequence of shorter date ranges. The
rationale is that by splitting the search into date ranges, users can
download multiple .csv files, which are then combined by the function
into a single metadata data frame. Of course users must download the csv
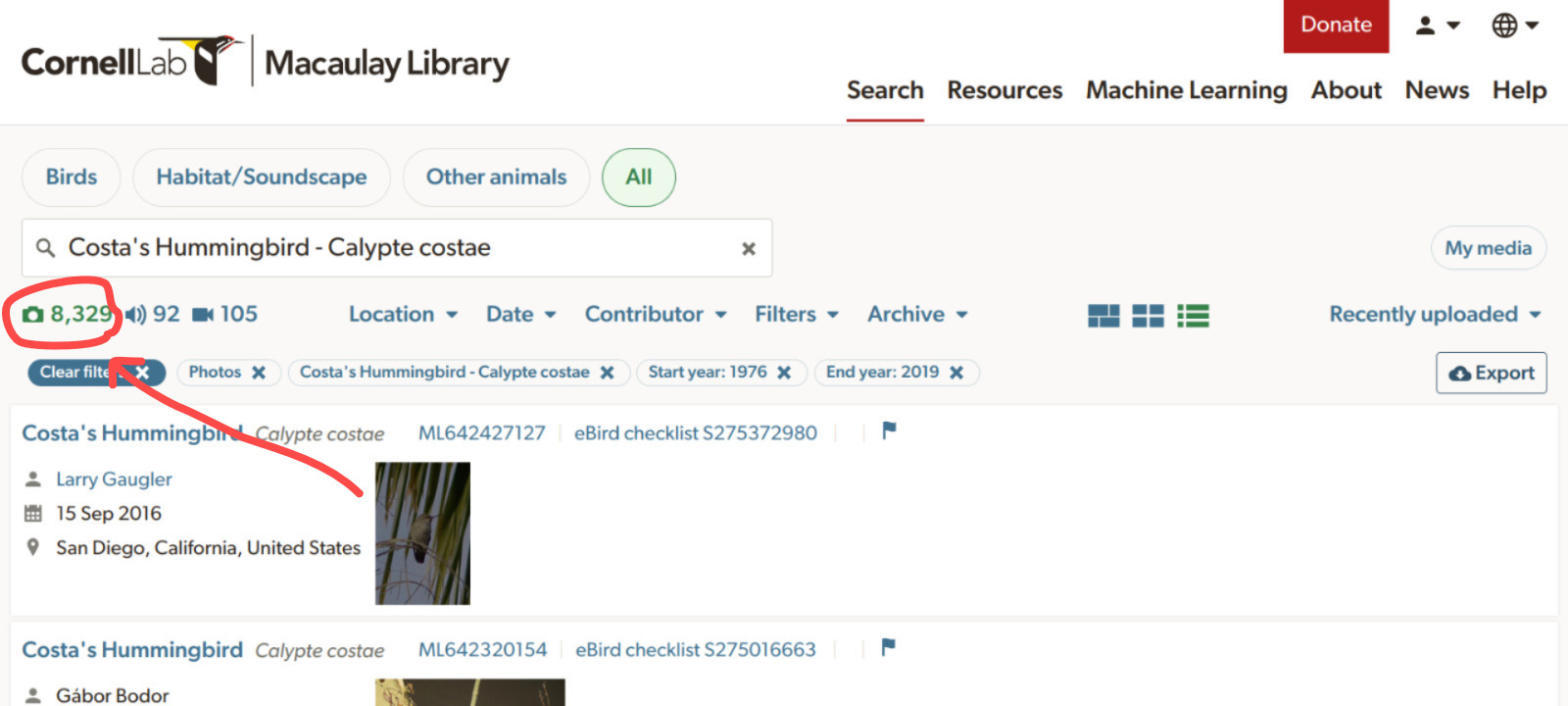
for each data range. The following code looks for photos of costa’s
hummingbird (Calypte costae). As Macaulay Library hosts more
than 30000 costa’s hummingbird records, we need to split the query into
multiple date ranges:
# test a query with more than 10000 results paging by date
cal_cos <- query_macaulay(
species = "Calypte costae",
format = "image",
path = tempdir(),
dates = c(1976, 2019, 2022, 2024, 2025, 2026)
)Users can check at the Macaulay Library website how many records are available for their species of interest (see image below) and then decide how to split the search by date ranges accordingly so each sub-query has less than 10000 records.
query_macaulay()
can also read metadata previously downloaded from Macaulay Library website. To
do this, users must provide 1) the name of the csv file(s) to the
argument files and 2) the directory path were it was saved
to the argument path.
query_xenocanto()
API key
Xeno-Canto requires users
to obtain a free API key to use their API v3.
Users can get their API key by creating an account at Xeno-Canto’s registering
page. Once users have their API key, they can set it as a variable
in your R environment using
Sys.setenv(xc_api_key = "YOUR_API_KEY_HERE") and query_xenocanto()
will use it. Here is an example of a query for Spix’s disc-winged bat
(Thyroptera tricolor) audio recordings:
# set your Xeno-Canto key as environmental variable
Sys.setenv(xc_api_key = "YOUR_API_KEY_HERE")
# query Xeno-CAnto
t_tricolor <- query_xenocanto(species = "Thyroptera tricolor")
head(t_tricolor, 4)| repository | format | key | species | date | time | user_name | country | locality | latitude | longitude | file_url | file_extension |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xeno-Canto | sound | 879621 | Thyroptera tricolor | 2023-07-15 | 12:30 | José Tinajero | Costa Rica | Hacienda Baru, Dominical, Costa Rica | 9.2635 | -83.8768 | https://xeno-canto.org/879621/download | wav |
| Xeno-Canto | sound | 820604 | Thyroptera tricolor | 2013-01-10 | 19:00 | Sébastien J. Puechmaille | Costa Rica | Pavo, Provincia de Puntarenas | 8.4815 | -83.5945 | https://xeno-canto.org/820604/download | wav |
| Xeno-Canto | sound | 820603 | Thyroptera tricolor | 2013-01-10 | 19:00 | Sébastien J. Puechmaille | Costa Rica | Pavo, Provincia de Puntarenas | 8.4815 | -83.5945 | https://xeno-canto.org/820603/download | wav |
| Xeno-Canto | sound | 821928 | Thyroptera tricolor | 2013-01-10 | 19:00 | Daniel j buckley | Costa Rica | Pavo, Provincia de Puntarenas | 8.4815 | -83.5945 | https://xeno-canto.org/821928/download | wav |
Special queries
query_xenocanto() allows users to perform special queries by specifying additional query tags. Users can also search by country, taxonomy (taxonomic group, family, genus, subspecies), geography (country, location, geographic coordinates) date, sound type (e.g. female song, calls) and recording properties (quality, length, sampling rate) (see list of available tags here). Here is an example of a query for audio recordings of pale-striped poison frog (Ameerega hahneli, ’sp:“Ameerega hahneli”) from French Guiana (cnt:“French Guiana”) and with the highest recording quality (q:“A”):
# assuming you already set your API key as in previous code block
a_hahneli <- query_xenocanto(
species = 'sp:"Ameerega hahneli" cnt:"French Guiana" q:"A"')
head(a_hahneli, 4)| repository | format | key | species | date | time | user_name | country | locality | latitude | longitude | file_url | file_extension |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xeno-Canto | sound | 928987 | Ameerega hahneli | 2024-05-14 | 16:00 | Augustin Bussac | French Guiana | Sentier Gros-Arbre | 3.6132 | -53.2169 | https://xeno-canto.org/928987/download | mp3 |
| Xeno-Canto | sound | 928972 | Ameerega hahneli | 2024-04-24 | 17:00 | Augustin Bussac | French Guiana | Camp Bonaventure | 4.3226 | -52.3387 | https://xeno-canto.org/928972/download | mp3 |
| Xeno-Canto | sound | 928971 | Ameerega hahneli | 2023-11-26 | 13:00 | Augustin Bussac | French Guiana | Guyane Natural Regional Park (near Roura), Arrondissement of Cayenne | 4.5423 | -52.4432 | https://xeno-canto.org/928971/download | mp3 |
Update metadata
The update_metadata()
function allows users to update a previous query to add new information
from the corresponding repository of the original search. This function
takes as input a data frame previously obtained from any query function
(i.e. query_reponame()) and returns a data frame similar to
the input with new data appended.
To show case the function, we first query metadata of Eisentraut’s Bow-winged Grasshopper sounds from iNaturalist. Let’s assume that the initial query was done a while ago and we want to update it to include any new records that might have been added since then. The following code removes all observations recorded after 2024-12-31 to simulate an old query:
# initial query
c_eisentrauti <- query_inaturalist(species = "Chorthippus eisentrauti")
[32m✔
[39m Obtaining metadata (113 matching records found) 🎊:
head(c_eisentrauti, 4)| repository | format | key | species | date | time | user_name | country | locality | latitude | longitude | file_url | file_extension |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| iNaturalist | image | 335245347 | Chorthippus eisentrauti | 2019-11-16 | 12:48 | Eliot Stein-Deffarges J. | NA | 43.967696755,7.6218244195 | 43.96770 | 7.621824 | https://inaturalist-open-data.s3.amazonaws.com/photos/608983424/original.jpg | jpeg |
| iNaturalist | image | 335245344 | Chorthippus eisentrauti | 2019-11-16 | 12:36 | Eliot Stein-Deffarges J. | NA | 43.967696755,7.6218244195 | 43.96770 | 7.621824 | https://inaturalist-open-data.s3.amazonaws.com/photos/608982971/original.jpg | jpeg |
| iNaturalist | image | 334597801 | Chorthippus eisentrauti | 2026-01-11 | 12:01 | Eliot Stein-Deffarges J. | NA | 44.0786166389,7.6128199722 | 44.07862 | 7.612820 | https://inaturalist-open-data.s3.amazonaws.com/photos/607665238/original.jpg | jpeg |
| iNaturalist | image | 332517118 | Chorthippus eisentrauti | 2023-08-13 | 13:07 | Parnassius | NA | 44.963801903,6.577495847 | 44.96380 | 6.577496 | https://inaturalist-open-data.s3.amazonaws.com/photos/603468000/original.jpg | jpeg |
# exclude new observations (simulate old data)
old_c_eisentrauti <-
c_eisentrauti[c_eisentrauti$date <= "2024-12-31" | is.na(c_eisentrauti$date),
]
# update "old" data
upd_c_eisentrauti <- update_metadata(metadata = old_c_eisentrauti)
[32m✔
[39m Obtaining metadata (113 matching records found) 🌈:
[32m✔
[39m 95 new entries found 🎊[1] FALSECombine metadata from multiple repositories
The merge_metadata()
function allows users to combine metadata data frames obtained from
multiple query functions into a single data frame. The function will
match the basic columns of all data frames. Data from additional columns
(for instance when using all_data = TRUE in the query) will
only be combined if the column names from different repositories match.
The function will return a data frame that includes a new column called
source indicating the name of the original metadata data
frame:
truf_xc <- query_xenocanto(species = "Turdus rufiventris",
format = "sound")
truf_gbf <- query_gbif(species = "Turdus rufiventris", format = "sound")
truf_ml <- query_macaulay(species = "Turdus rufiventris",
format = "sound",
path = tempdir())
# merge metadata
merged_metadata <- merge_metadata(truf_xc, truf_gbf, truf_ml)
head(merged_metadata, 4)| repository | format | key | species | date | time | user_name | country | locality | latitude | longitude | file_url | file_extension | source |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xeno-Canto | sound | 1032061 | Turdus rufiventris | 2025-07-19 | 18:01 | Jacob Wijpkema | Bolivia | Lagunillas, Cordillera, Santa Cruz Department | -19.6348 | -63.6711 | https://xeno-canto.org/1032061/download | wav | xc_adf |
| Xeno-Canto | sound | 1006659 | Turdus rufiventris | 2025-06-13 | 16:01 | Jayrson Araujo De Oliveira | Brazil | RPPN Flor das Águas - Pirenópolis, Goiás | -15.8195 | -48.9861 | https://xeno-canto.org/1006659/download | mp3 | xc_adf |
| Xeno-Canto | sound | 979639 | Turdus rufiventris | 2025-03-12 | 08:00 | Ricardo José Mitidieri | Brazil | Tijuca, Teresópolis, Rio de Janeiro | -22.4212 | -42.9559 | https://xeno-canto.org/979639/download | mp3 | xc_adf |
| Xeno-Canto | sound | 974643 | Turdus rufiventris | 2025-02-14 | 13:11 | Jayrson Araujo De Oliveira | Brazil | Fazenda Nazareth Eco, Jose de Freitas-PI, Piauí | -4.7958 | -42.6150 | https://xeno-canto.org/974643/download | mp3 | xc_adf |
Note that in such a multi-repository query, all query functions use
the same search species (i.e. species name) and media format (e.g.,
sound, image, video). To facilitate this, users can set the global
options species and format so they do not need
to specify them in each query function:
options(species = "Turdus rufiventris", format = "sound")
truf_xc <- query_xenocanto() # assuming you already set your API key
truf_gbf <- query_gbif()
truf_ml <- query_macaulay(path = tempdir())
# merge metadata
merged_metadata <- merge_metadata(truf_xc, truf_gbf, truf_ml)Find and remove duplicated records
When compiling data from multiple repositories, duplicated media records are a common issue, particularly for sound recordings. These duplicates occur both through data sharing between repositories like Xeno-Canto and GBIF, and when users upload the same file to multiple platforms. To help users efficiently identify these duplicate records, suwo provides the find_duplicates() function. Duplicates are identified based on matching species name, country, date, user name, and locality. The function uses a fuzzy matching approach to account for minor variations in the data (e.g., typos, different location formats, etc).The output is a data frame with the candidate duplicate records, allowing users to review and decide which records to keep.
In this example we look for possible duplicates in the merged metadata data frame from the previous section:
# find duplicates
dups_merged_metadata <- find_duplicates(merged_metadata)
[36mℹ
[39m 611 potential duplicates found
# look first 6 columns
head(dups_merged_metadata)| repository | format | key | species | date | time | user_name | country | locality | latitude | longitude | file_url | file_extension | source | duplicate_group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xeno-Canto | sound | 913487 | Turdus rufiventris | 2024-06-07 | 06:46 | Jayrson Araujo De Oliveira | Brazil | Reserva do Setor Sítio de Recreio Caraíbas-Goiânia, Goiás | -16.5631 | -49.2850 | https://xeno-canto.org/913487/download | mp3 | xc_adf | 1 |
| GBIF | sound | 4907346188 | Turdus rufiventris | 2024-06-07 | 06:46 | Jayrson Araujo De Oliveira | Brazil | Reserva do Setor Sítio de Recreio Caraíbas-Goiânia, Goiás | -16.5631 | -49.2850 | https://xeno-canto.org/sounds/uploaded/LXKLWEDKEM/XC913487-07-06-2024-6e46-Sabia-laranjeira-CARAIBAS.mp3 | mp3 | gb_adf_s | 1 |
| Xeno-Canto | sound | 351258 | Turdus rufiventris | 2013-10-11 | 17:27 | Jeremy Minns | Brazil | Miranda, MS. Refúgio da Ilha | -20.2209 | -56.5751 | https://xeno-canto.org/351258/download | mp3 | xc_adf | 2 |
| GBIF | sound | 2243749719 | Turdus rufiventris | 2013-10-11 | 17:27 | Jeremy Minns | Brazil | Miranda, MS. Refúgio da Ilha | -20.2209 | -56.5751 | https://xeno-canto.org/sounds/uploaded/DGVLLRYDXS/XC351258-TURRUF68.mp3 | mp3 | gb_adf_s | 2 |
| Xeno-Canto | sound | 351066 | Turdus rufiventris | 2013-10-10 | 17:02 | Jeremy Minns | Brazil | Miranda, MS. Refúgio da Ilha | -20.2209 | -56.5751 | https://xeno-canto.org/351066/download | mp3 | xc_adf | 3 |
| GBIF | sound | 2243747991 | Turdus rufiventris | 2013-10-10 | 17:02 | Jeremy Minns | Brazil | Miranda, MS. Refúgio da Ilha | -20.2209 | -56.5751 | https://xeno-canto.org/sounds/uploaded/DGVLLRYDXS/XC351066-TURRUF67.mp3 | mp3 | gb_adf_s | 3 |
Note that the find_duplicates() function adds a new column called “duplicate_group” to the output data frame. This column assigns a unique identifier to each group of potential duplicates, allowing users to easily identify and review them. For instance, in the example above, records from duplicated group 75 belong to the same user, were recorded on the same date and time and in the same country:
subset(dups_merged_metadata, duplicate_group == 75)| repository | format | key | species | date | time | user_name | country | locality | latitude | longitude | file_url | file_extension | source | duplicate_group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xeno-Canto | sound | 273100 | Turdus rufiventris | 2013-10-19 | 18:00 | Peter Boesman | Argentina | Calilegua NP, Jujuy | -23.74195 | -64.85777 | https://xeno-canto.org/273100/download | mp3 | xc_adf | 75 |
| Xeno-Canto | sound | 273098 | Turdus rufiventris | 2013-10-19 | 18:00 | Peter Boesman | Argentina | Calilegua NP, Jujuy | -23.74195 | -64.85777 | https://xeno-canto.org/273098/download | mp3 | xc_adf | 75 |
| GBIF | sound | 2243678570 | Turdus rufiventris | 2013-10-19 | 18:00 | Peter Boesman | Argentina | Calilegua NP, Jujuy | -23.74195 | -64.85777 | https://xeno-canto.org/sounds/uploaded/OOECIWCSWV/XC273098-Rufous-bellied%20Thrush%20QQ%20call%20A%201.mp3 | mp3 | gb_adf_s | 75 |
| GBIF | sound | 2243680322 | Turdus rufiventris | 2013-10-19 | 18:00 | Peter Boesman | Argentina | Calilegua NP, Jujuy | -23.74195 | -64.85777 | https://xeno-canto.org/sounds/uploaded/OOECIWCSWV/XC273100-Rufous-bellied%20Thrush%20QQQ%20call%20A.mp3 | mp3 | gb_adf_s | 75 |
| Macaulay Library | sound | 301276 | Turdus rufiventris | 2013-10-19 | 18:00 | Peter Boesman | Argentina | Calilegua NP | -23.74200 | -64.85780 | https://cdn.download.ams.birds.cornell.edu/api/v1/asset/301276/ | mp3 | ml_adf_s | 75 |
| Macaulay Library | sound | 301275 | Turdus rufiventris | 2013-10-19 | 18:00 | Peter Boesman | Argentina | Calilegua NP | -23.74200 | -64.85780 | https://cdn.download.ams.birds.cornell.edu/api/v1/asset/301275/ | mp3 | ml_adf_s | 75 |
Also note that the locality is not exactly the same for these
records, but the fuzzy matching approach used by find_duplicates()
was able to identify them as potential duplicates. By default, the
criteria is set to
country > 0.8 & locality > 0.5 & user_name > 0.8 & time == 1 & date == 1
which means that two entries will be considered duplicates if they have
a country similarity greater than 0.8, locality similarity greater than
0.5, user_name similarity greater than 0.8, and exact matches for time
and date (similarities range from 0 to 1). These values have been found
to work well in most cases. Nonetheless, users can adjust the
sensitivity based on their specific needs using the argument
criteria.
Once users have reviewed the candidate duplicates, they can apply the remove_duplicates() function to eliminate unwanted duplicates from their metadata data frames. This function takes as input a metadata data frame (either the original query results or the output of find_duplicates()) and a vector of row numbers indicating which records to remove:
# remove duplicates
dedup_metadata <- remove_duplicates(dups_merged_metadata)
[36mℹ
[39m 263 duplicates removed The output is a data frame similar to the input but without the specified duplicate records:
# look at first 4 columns of deduplicated metadata
head(dedup_metadata, 4)| repository | format | key | species | date | time | user_name | country | locality | latitude | longitude | file_url | file_extension | source | duplicate_group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xeno-Canto | sound | 913487 | Turdus rufiventris | 2024-06-07 | 06:46 | Jayrson Araujo De Oliveira | Brazil | Reserva do Setor Sítio de Recreio Caraíbas-Goiânia, Goiás | -16.5631 | -49.2850 | https://xeno-canto.org/913487/download | mp3 | xc_adf | 1 |
| GBIF | sound | 4907346188 | Turdus rufiventris | 2024-06-07 | 06:46 | Jayrson Araujo De Oliveira | Brazil | Reserva do Setor Sítio de Recreio Caraíbas-Goiânia, Goiás | -16.5631 | -49.2850 | https://xeno-canto.org/sounds/uploaded/LXKLWEDKEM/XC913487-07-06-2024-6e46-Sabia-laranjeira-CARAIBAS.mp3 | mp3 | gb_adf_s | 1 |
| Xeno-Canto | sound | 351258 | Turdus rufiventris | 2013-10-11 | 17:27 | Jeremy Minns | Brazil | Miranda, MS. Refúgio da Ilha | -20.2209 | -56.5751 | https://xeno-canto.org/351258/download | mp3 | xc_adf | 2 |
| GBIF | sound | 2243749719 | Turdus rufiventris | 2013-10-11 | 17:27 | Jeremy Minns | Brazil | Miranda, MS. Refúgio da Ilha | -20.2209 | -56.5751 | https://xeno-canto.org/sounds/uploaded/DGVLLRYDXS/XC351258-TURRUF68.mp3 | mp3 | gb_adf_s | 2 |
When duplicates are found, one observation from each group of
duplicates is retained in the output data frame. However, if multiple
observations from the same repository are labeled as duplicates, by
default (same_repo = FALSE) all of them are retained in the
output data frame. This is useful as it can be expected that
observations from the same repository are not true duplicates
(e.g. different recordings uploaded to Xeno-Canto with the same date,
time and location by the same user), but rather have not been documented
with enough precision to be told apart. This behavior can be modified.
If same_repo = TRUE, only one of the duplicated
observations from the same repository will be retained in the output
data frame (and all other excluded). The function will give priority to
repositories in which media downloading is more straightforward
(i.e. Xeno-Canto, GBIF), but this can be modified with the argument
repo_priority.
Download media files
The last step of the workflow is to download the media files associated with the metadata. This can be done using the download_media() function, which takes as input a metadata data frame (obtained from any query function or any of the other metadata managing functions) and downloads the media files to a specified directory. For this example we will download images from a query on zambian slender Caesar (Amanita zambiana) (a mushroom) on GBIF:
# query GBIF for Amanita zambiana images
a_zam <- query_gbif(species = "Amanita zambiana", format = "image")
[32m✔
[39m Obtaining metadata (7 matching records found) 🥇:
# create folder for images
out_folder <- file.path(tempdir(), "amanita_zambiana")
dir.create(out_folder)
# download media files to a temporary directory
azam_files <- download_media(metadata = a_zam, path = out_folder)Downloading media files:
[32m✔
[39m All files were downloaded successfully 🎉The output of the function is a data frame similar to the input metadata but with two additional columns indicating the file name of the downloaded files (‘downloaded_file_name’) and the result of the download attempt (‘download_status’, with values “success”, ‘failed’, ‘already there (not downloaded)’ or ‘overwritten’).
Here we print the first 4 rows of the output data frame:
head(azam_files, 4)| repository | format | key | species | date | time | user_name | country | locality | latitude | longitude | file_url | file_extension | downloaded_file_name | download_status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GBIF | image | 4430877067 | Amanita zambiana | 2023-01-25 | 10:57 | Allanweideman | Mozambique | NA | -21.28456 | 34.61868 | https://inaturalist-open-data.s3.amazonaws.com/photos/253482452/original.jpg | jpeg | Amanita_zambiana-GBIF4430877067-1.jpeg | saved |
| GBIF | image | 4430877067 | Amanita zambiana | 2023-01-25 | 10:57 | Allanweideman | Mozambique | NA | -21.28456 | 34.61868 | https://inaturalist-open-data.s3.amazonaws.com/photos/253482473/original.jpg | jpeg | Amanita_zambiana-GBIF4430877067-2.jpeg | saved |
| GBIF | image | 4430877067 | Amanita zambiana | 2023-01-25 | 10:57 | Allanweideman | Mozambique | NA | -21.28456 | 34.61868 | https://inaturalist-open-data.s3.amazonaws.com/photos/253484256/original.jpg | jpeg | Amanita_zambiana-GBIF4430877067-3.jpeg | saved |
| GBIF | image | 5104283819 | Amanita zambiana | 2023-03-31 | 13:41 | Nick Helme | Zambia | NA | -12.44276 | 31.28535 | https://inaturalist-open-data.s3.amazonaws.com/photos/268158445/original.jpeg | jpeg | Amanita_zambiana-GBIF5104283819.jpeg | saved |
… and check that the files were saved in the path supplied:
fs::dir_tree(path = out_folder)
/tmp/Rtmpw4Mwx9/amanita_zambiana
├──
Amanita_zambiana-GBIF3759537817-1.jpeg
├──
Amanita_zambiana-GBIF3759537817-2.jpeg
├──
Amanita_zambiana-GBIF4430877067-1.jpeg
├──
Amanita_zambiana-GBIF4430877067-2.jpeg
├──
Amanita_zambiana-GBIF4430877067-3.jpeg
├──
Amanita_zambiana-GBIF5069132689-1.jpeg
├──
Amanita_zambiana-GBIF5069132689-2.jpeg
├──
Amanita_zambiana-GBIF5069132691.jpeg
├──
Amanita_zambiana-GBIF5069132696-1.jpeg
├──
Amanita_zambiana-GBIF5069132696-2.jpeg
├──
Amanita_zambiana-GBIF5069132732.jpeg
└──
Amanita_zambiana-GBIF5104283819.jpeg
Note that the name of the downloaded files includes the species name, an abbreviation of the repository name and the unique record key. If more than one media file is associated with a record, a sequential number is added at the end of the file name.
This is a multipanel plot of 6 of the downloaded images (just for illustration purpose):
# create a 6 pannel plot of the downloaded images
opar <- par(mfrow = c(2, 3), mar = c(1, 1, 2, 1))
for (i in 1:6) {
img <- jpeg::readJPEG(file.path(out_folder, azam_files$downloaded_file_name[i]))
plot(
1:2,
type = 'n',
axes = FALSE
)
graphics::rasterImage(img, 1, 1, 2, 2)
title(main = paste(
azam_files$country[i],
azam_files$date[i],
sep = "\n"
))
}
# reset par
par(opar)Users can also save the downloaded files into sub-directories with
the argument folder_by. This argument takes a character or
factor column with the names of a metadata field (a column in the
metadata data frame) to create sub-directories within the main download
directory (suplied with the argument path). For instance,
the following code searches/downloads images of longspined porcupinefish
(Diodon holocanthus) from GBIF, and saves images into
sub-directories by country (for simplicity only 6 of them):
# query GBIF for longspined porcupinefish images
d_holocanthus <- query_gbif(species = "Diodon holocanthus", format = "image")
# keep only JPEG records (for simplicity for this vignette)
d_holocanthus <- d_holocanthus[d_holocanthus$file_extension == "jpeg", ]
# select 6 random JPEG records
set.seed(666)
d_holocanthus <- d_holocanthus[sample(seq_len(nrow(d_holocanthus)), 6),]
# create folder for images
out_folder <- file.path(tempdir(), "diodon_holocanthus")
dir.create(out_folder)
# download media files creating sub-directories by country
dhol_files <- download_media(metadata = d_holocanthus,
path = out_folder,
folder_by = "country")Downloading media files:
[32m✔
[39m All files were downloaded successfully 🎊
fs::dir_tree(path = out_folder)
/tmp/Rtmpw4Mwx9/diodon_holocanthus
├──
Cabo Verde
│ └──
Diodon_holocanthus-GBIF3985886532.jpeg
├──
Cayman Islands
│ └──
Diodon_holocanthus-GBIF5827468492.jpeg
├──
Chinese Taipei
│ └──
Diodon_holocanthus-GBIF5206745484.jpeg
├──
Indonesia
│ └──
Diodon_holocanthus-GBIF4953086522.jpeg
└──
United States of America
├──
Diodon_holocanthus-GBIF1270050026.jpeg
└──
Diodon_holocanthus-GBIF4935688405.jpeg
In such case the ‘downloaded_file_name’ column will include the sub-directory name:
dhol_files$downloaded_file_name[1] "United States of America/Diodon_holocanthus-GBIF4935688405.jpeg"
[2] "United States of America/Diodon_holocanthus-GBIF1270050026.jpeg"
[3] "Cabo Verde/Diodon_holocanthus-GBIF3985886532.jpeg"
[4] "Cayman Islands/Diodon_holocanthus-GBIF5827468492.jpeg"
[5] "Chinese Taipei/Diodon_holocanthus-GBIF5206745484.jpeg"
[6] "Indonesia/Diodon_holocanthus-GBIF4953086522.jpeg" This is a multipanel plot of the downloaded images (just for fun):
# create a 6 pannel plot of the downloaded images
opar <- par(mfrow = c(2, 3), mar = c(1, 1, 2, 1))
for (i in 1:6) {
img <- jpeg::readJPEG(file.path(out_folder, dhol_files$downloaded_file_name[i]))
plot(
1:2,
type = 'n',
axes = FALSE
)
graphics::rasterImage(img, 1, 1, 2, 2)
title(main = paste(
substr(dhol_files$country[i], start = 1, stop = 14),
dhol_files$date[i],
sep = "\n"
))
}
# reset par
par(opar)Session information
Click to see
R version 4.5.2 (2025-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 24.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8 LC_COLLATE=C.UTF-8
[5] LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8 LC_PAPER=C.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
time zone: UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] kableExtra_1.4.0 suwo_0.1.0 knitr_1.51
loaded via a namespace (and not attached):
[1] viridisLite_0.4.3 farver_2.1.2 blob_1.3.0 fastmap_1.2.0
[5] digest_0.6.39 rpart_4.1.24 timechange_0.4.0 lifecycle_1.0.5
[9] survival_3.8-3 RSQLite_2.4.5 magrittr_2.0.4 compiler_4.5.2
[13] rlang_1.1.7 sass_0.4.10 tools_4.5.2 yaml_2.3.12
[17] data.table_1.18.2.1 htmlwidgets_1.6.4 bit_4.6.0 curl_7.0.0
[21] xml2_1.5.2 RColorBrewer_1.1-3 desc_1.4.3 nnet_7.3-20
[25] grid_4.5.2 xtable_1.8-4 e1071_1.7-17 future_1.69.0
[29] globals_0.19.0 scales_1.4.0 MASS_7.3-65 cli_3.6.5
[33] rmarkdown_2.30 crayon_1.5.3 ragg_1.5.0 generics_0.1.4
[37] rstudioapi_0.18.0 RecordLinkage_0.4-12.6 future.apply_1.20.1 DBI_1.2.3
[41] pbapply_1.7-4 cachem_1.1.0 proxy_0.4-29 stringr_1.6.0
[45] splines_4.5.2 parallel_4.5.2 vctrs_0.7.1 Matrix_1.7-4
[49] jsonlite_2.0.0 bit64_4.6.0-1 listenv_0.10.0 jpeg_0.1-11
[53] systemfonts_1.3.1 evd_2.3-7.1 jquerylib_0.1.4 glue_1.8.0
[57] parallelly_1.46.1 pkgdown_2.2.0 codetools_0.2-20 lubridate_1.9.5
[61] stringi_1.8.7 tibble_3.3.1 pillar_1.11.1 rappdirs_0.3.4
[65] htmltools_0.5.9 ipred_0.9-15 lava_1.8.2 R6_2.6.1
[69] ff_4.5.2 httr2_1.2.2 textshaping_1.0.4 evaluate_1.0.5
[73] lattice_0.22-7 backports_1.5.0 memoise_2.0.1 bslib_0.10.0
[77] class_7.3-23 Rcpp_1.1.1 svglite_2.2.2 prodlim_2025.04.28
[81] checkmate_2.3.4 xfun_0.56 fs_1.6.6 pkgconfig_2.0.3