Visual inspection and signal classification
Marcelo Araya-Salas, PhD & Grace Smith-Vidaurre
2024-08-19
Source:vignettes/warbleR_workflow_02.Rmd
warbleR_workflow_02.RmdBioacoustics in R with warbleR

Bioacoustics research encompasses a wide range of questions, study
systems and methods, including the software used for analyses. The
warbleR and Rraven packages leverage the
flexibility of the R environment to offer a broad and
accessible bioinformatics tool set. These packages fundamentally rely
upon two types of data to begin bioacoustic analyses in R:
Sound files: Recordings in wav or mp3 format, either from your own research or open-access databases like xeno-canto
Selection tables: Selection tables contain the temporal coordinates (start and end points) of selected acoustic signals within recordings
Package repositories
These packages are both available on CRAN: warbleR,
Rraven,
as well as on GitHub: warbleR, Rraven. The
GitHub repository will always contain the latest functions and updates.
You can also check out an article in Methods in Ecology and
Evolution documenting the warbleR package
[1].
We welcome all users to provide feedback, contribute updates or new functions and report bugs to warbleR’s GitHub repository.
Please note that warbleR and Rraven use
functions from the seewave,
monitoR,
tuneR
and dtw
packages internally. warbleR and Rraven have
been designed to make bioacoustics analyses more accessible to
R users, and such analyses would not be possible without
the tools provided by the packages above. These packages should be given
credit when using warbleR and Rraven by
including citations in publications as appropriate
(e.g. citation("seewave")).
Parallel processing in warbleR
Parallel processing, or using multiple cores on your machine, can
greatly speed up analyses. All iterative warbleR functions
now have parallel processing for Linux, Mac and Windows operating
systems. These functions also contain progress bars to visualize
progress during normal or parallel processing. See
[1] for more details about improved
running time using parallel processing.
Vignette introduction
In the previous vignette, we used the Rraven package to
import Raven selection tables for recordings in our working
directory, added more recordings to the data set by downloading new
sound files from the open-access xeno-canto database and
reviewed methods of automated and manual signal selection in
warbleR. Here we continue with the case study of
microgeographic vocal variation in long-billed hermit hummingbirds,
Phaethornis longirostris
[2] by:
Performing quality control processing on selected signals, including visual inspection and tailoring temporal coordinates
Making lexicons for visual classification of signals
This vignette can be run without an advanced understanding of
R, as long as you know how to run code in your console.
However, knowing more about basic R coding would be very
helpful to modify the code for your research questions.
For more details about function arguments, input or output, read the
documentation for the function in question (e.g. ?catalog).
Prepare for analyses
library(warbleR)
# set your working directory appropriately
# setwd("/path/to/working directory")
# run this if you have restarted RStudio between vignettes without saving your workspace (assuming that you are in your /home/username directory)
setwd(file.path(getwd(), "warbleR_example"))
# Check your location
getwd()This vignette series will not always include all available
warbleR functions, as existing functions are updated and
new functions are added. To see all functions available in this
package:
# The package must be loaded in your working environment
ls("package:warbleR")Quality control filtering of selections
Find overlapping selections
Overlapping selections can sometimes arise after selecting signals using other functions or software. The function below helps you detect overlapping signals in your selection table, and has arguments that you can play around with for overlap detection, renaming or deleting overlapping selections.
# To run this example:
# Open Phae_hisnr.csv and modify the start coordinate of the first selection and the end coordinate of the second selection so that the signals overlap
Phae.hisnr <- read.csv("Phae_hisnr.csv", header = TRUE)
str(Phae.hisnr)
head(Phae.hisnr, n = 15)
# yields a data frame with an additional column (ovlp.sels) that indicates which selections overlap
Phae.hisnr <- overlapping_sels(X = Phae.hisnr, max.ovlp = 0)
# run the function again but this time retain only the signals that don't overlap
Phae.hisnr <- overlapping_sels(X = Phae.hisnr, max.ovlp = 0, drop = TRUE)Make spectrograms of selections
spectrograms generates spectrograms of individual
selected signals. These image files can be used to filter out selections
that were poorly made or represent signals that are not relevant to your
analysis. This quality control step is important for visualizing your
selected signals after any selection method, even if you imported your
selections from Raven or Syrinx.
spectrograms(Phae.hisnr, wl = 300, flim = c(2, 10), it = "jpeg", res = 150, osci = TRUE, ovlp = 90)Inspect spectrograms and throw away image files that are poor quality to prepare for later steps. Make sure you are working in a directory that only has image files associated with this vignette. Delete the image files corresponding to recording 154070 selection 8 and 154070 selection 12, as the start coordinates for these selections are not accurate.
Remove selections with missing image files
# remove selections after deleting corresponding image files
Phae.hisnr2 <- filter_sels(Phae.hisnr, it = "jpeg", incl.wav = TRUE)
nrow(Phae.hisnr2)After removing the poorest quality selections or signals, there are some other quality control steps that may be helpful.
Check selections
Can selections be read by downstream functions? The function
checksels also yields a data frame with columns for
duration, minimum samples, sampling rate, channels and bits.
# if selections can be read, "OK" will be printed to check.res column
checksels(Phae.hisnr2, check.header = FALSE)If selections cannot be read, it is possible the sound files are
corrupt. If so, use the fixwavs function to repair
wav files.
Tailor temporal coordinates of selections
Sometimes the start and end times of selected signals need fine-tuned
adjustments. This is particularly true when signals are found within
bouts of closely delivered sounds that may be hard to pull apart, such
as duets, or if multiple researchers use different rules-of-thumb to
select signals. tailor_sels provides an interactive
interface for tailoring the temporal coordinates of selections.
If you check out the image files generated by running
spectrograms above, you’ll see that some of the selections
made during the automatic detection process with auto_detec
do not have accurate start and/or end coordinates.
For instance:
The end of this signal is not well selected.
The temporal coordinates for the tailored signals will be saved in a
_ .csv_ file called seltailor_output.csv. You can rename
this file and read it back into R to continue downstream
analyses.
tailor_sels(Phae.hisnr2, wl = 300, flim = c(2, 10), wn = "hanning", mar = 0.1, osci = TRUE, title = c("sound.files", "selec"), auto.next = TRUE)
# Read in tailor_sels output after renaming the csv file
Phae.hisnrt <- read.csv("Phae_hisnrt.csv", header = TRUE)
str(Phae.hisnrt)'data.frame': 23 obs. of 6 variables:
$ sound.files: chr "Phaethornis-longirostris-154070.wav" "Phaethornis-longirostris-154070.wav" "Phaethornis-longirostris-154070.wav" "Phaethornis-longirostris-154072.wav" ...
$ selec : int 14 18 20 36 108 124 142 148 57 83 ...
$ start : num 8.66 11.08 12.19 22.6 76.04 ...
$ end : num 8.79 11.2 12.32 22.73 76.16 ...
$ SNR : num 11.7 11.6 10.2 15.5 15.3 ...
$ tailored : chr "y" "y" "y" "y" ...Visual classification of selected signals
Visual classification of signals is fundamental to vocal repertoire
analysis, and can also be useful for other questions. If your research
focuses on assessing variation between individuals or groups, several
warbleR functions can provide you with important
information about how to steer your analysis. If there is discrete
variation in vocalization structure across groups (e.g. treatments or
geographic regions), visual classification of vocalizations will be
useful.
Print long spectrograms with full_spectrograms
The function full_spectrogramsthat we used in the last
vignette can also be a tool for visually classifying signals. Long
spectrograms can be printed to classify signals by hand, or comments
accompanying the selections can be printed over selected signals.
Here, we print the start and end of selections with a red dotted line, and the selection number printed over the signal. If a selection data frame contains a comments column, these will be printed with the selection number.
# highlight selected signals
full_spectrograms(Phae.hisnrt, wl = 300, flim = c(2, 10), ovlp = 10, sxrow = 6, rows = 15, it = "jpeg")
# concatenate full_spectrograms image files into a single PDF per recording
# full_spectrograms images must be jpegs
full_spectrograms2pdf(keep.img = FALSE, overwrite = TRUE)Check out the image file in your working directory. These will look
very similar to the full_spectrograms images produced in
vignette 1, but with red dotted lines indicating where the selected
signals start and end.
Highlight spectrogram regions with color_spectro
color_spectro allows you to highlight selections you’ve
made within a short region of a spectrogram. In the example below we
will use color_spectro to highlight neighboring songs. This
function has a wide variety of uses, and could be especially useful for
analysis of duets or coordinated singing bouts. This example is taken
directly from the color_spectro documentation. If working
with your own data frame of selections, make sure to calculate the
frequency range for your selections beforehand using the function
frange, which will come up in the next vignette.
# we will use Phaethornis songs and selections from the warbleR package
data(list = c("Phae.long1", "lbh_selec_table"))
writeWave(Phae.long1, "Phae.long1.wav") # save sound files
# subset selection table
# already contains the frequency range for these signals
st <- lbh_selec_table[lbh_selec_table$sound.files == "Phae.long1.wav", ]
# read wave file as an R object
sgnl <- read_sound_file(as.character(st$sound.files[1]))
# create color column
st$colors <- c("red2", "blue", "green")
# highlight selections
color_spectro(wave = sgnl, wl = 300, ovlp = 90, flim = c(1, 8.6), collevels = seq(-90, 0, 5), dB = "B", X = st, col.clm = "colors", base.col = "skyblue", t.mar = 0.07, f.mar = 0.1)Optimize spectrogram display parameters
spec_param makes a catalog or mosaic of the same signal
plotted with different combinations of spectrogram display arguments.
The purpose of this function is to help you choose parameters that yield
the best spectrograms (e.g. optimal visualization) for your signals
(although low signal-to-noise ratio selections may be an exception).
Make lexicons of signals
When we are interested in geographic variation of acoustic signals,
we usually want to compare spectrograms from different individuals and
sites. This can be challenging when working with large numbers of
signals, individuals and/or sites. catalog aims to simplify
this task.
This is how it works:
-
catalogplots a matrix of spectrograms from signals listed in a selection table - the catalog files are saved as image files in the working directory (or path provided)
- Several image files are generated if the signals do not fit in a single file
- Spectrograms can be labeled or color-tagged to facilitate exploring variation related to the parameter of interest (e.g. site or song type if already classified)
- A legend can be added to help match colors with tag levels
- different color palettes can be used for each tag
- The duration of the signals can be “fixed” such that all the
spectrograms have the same duration
- facilitates comparisons
- You can control the number of rows and columns as well as the width and height of the output image
catalog allows you to group signals into biologically
relevant groups by coloring the background of selected spectrograms
accordingly. There is also an option to add hatching to tag labels, as
well as filling the catalog with spectrograms by rows or columns of the
selection table data frame, among other additional arguments.
The move_imgs function can come in handy when creating
multiple catalogs to avoid overwriting previous image files, or when
working through rounds of other image files. In this case, the first
catalog we create has signals labeled, tagged and grouped with
respective color and hatching levels. The second catalog we create will
not have any grouping of signals whatsoever, and could be used for a
test of inter-observer reliability. move_imgs helps us move
the first catalog into another directory to save it from being
overwritten when creating the second catalog.
# create a column of recording IDs for friendlier catalog labels
rec_ID <- sapply(1:nrow(Phae.hisnrt), function(x) {
gsub(x = strsplit(as.character(Phae.hisnrt$sound.files[x]), split = "-")[[1]][3], pattern = ".wav$", replacement = "")
})
rec_ID
Phae.hisnrt$rec_ID <- rec_ID
str(Phae.hisnrt)
# set color palette
# alpha controls transparency for softer colors
cmc <- function(n) cm.colors(n, alpha = 0.8)
catalog(X = Phae.hisnrt, flim = c(2, 10), nrow = 4, ncol = 3, height = 10, width = 10, tag.pal = list(cmc), cex = 0.8, same.time.scale = TRUE, mar = 0.01, wl = 300, gr = FALSE, labels = "rec_ID", tags = "rec_ID", hatching = 1, group.tag = "rec_ID", spec.mar = 0.4, lab.mar = 0.8, max.group.cols = 5)
catalog2pdf(keep.img = FALSE, overwrite = TRUE)
# assuming we are working from the warbleR_example directory
# the ~/ format does not apply to Windows
# make sure you have already moved or deleted all other pdf files
move_images(from = ".", it = "pdf", create.folder = TRUE, folder.name = "Catalog_image_files")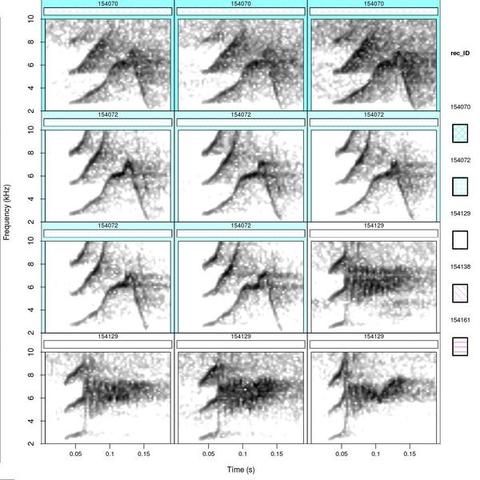
You can also make lexicons for blind scoring, which could be useful for determining interobserver reliability.
# now create a catalog without labels, tags, groups or axes
Phae.hisnrt$no_label <- ""
# catalog(X = Phae.hisnrt, flim = c(1, 10), nrow = 4, ncol = 3, height = 10, width = 10, cex = 0.8, same.time.scale = TRUE, mar = 0.01, wl = 300, spec.mar = 0.4, rm.axes = TRUE, labels = "no_label", lab.mar = 0.8, max.group.cols = 5, img.suffix = "nolabel")
catalog(X = Phae.hisnrt, flim = c(1, 10), nrow = 4, ncol = 3, height = 10, width = 10, tag.pal = list(cmc), cex = 0.8, same.time.scale = TRUE, mar = 0.01, wl = 300, gr = FALSE, labels = "no_label", spec.mar = 0.4, lab.mar = 0.8, max.group.cols = 5, img.suffix = "nolabels")
catalog2pdf(keep.img = FALSE, overwrite = TRUE)Next vignette: Acoustic (dis)similarity, coordinated singing and simulating songs
Here we finished the second phase of the warbleR
workflow, which includes various options for quality control filtering
or visual classification of signals that you can leverage during
acoustic analysis. After running the code in this vignette, you should
now have an idea of how to:
- how to perform quality control filtering of your selected signals, including visual inspection and tailoring temporal coordinates of spectrograms
- use different methods for visual classification of signals,
including:
- long spectrograms
- highlighted regions within spectrograms
- catalogs or lexicons of individual signals
The next vignette will cover the third phase of the warbleR workflow, which includes methods to perform acoustic measurements as a batch process, an example of how to use these measurements for an analysis of geographic variation, coordinated singing analysis and a new function to simulate songs.
Citation
Please cite warbleR when you use the package:
Araya-Salas, M. and Smith-Vidaurre, G. (2017), warbleR: an R package to streamline analysis of animal acoustic signals. Methods Ecol Evol. 8, 184-191.
Reporting bugs
Please report any bugs here.
- Araya-Salas, M. and G. Smith-Vidaurre. 2016. warbleR: an R package to streamline analysis of animal acoustic signals. Methods in Ecology and Evolution. doi: 10.1111/2041-210X.12624