R basics
Tropical Biology 2022
Organization for
Tropical Studies
Organization for Tropical Studies
Marcelo
Araya-Salas, PhD
“2022-06-15”
What is R?
- A free Domain-Specific-Language (DSL) for statistics and data analysis
- A collection of over 18949 (mar-9-2022) libraries
- A large and active community across industry and academia
- A way to talk “directly” to your computer
Historically:
- Based on the S Programming Language
- Around 20 years old (Lineage dates from 1975 - almost 40 years ago)
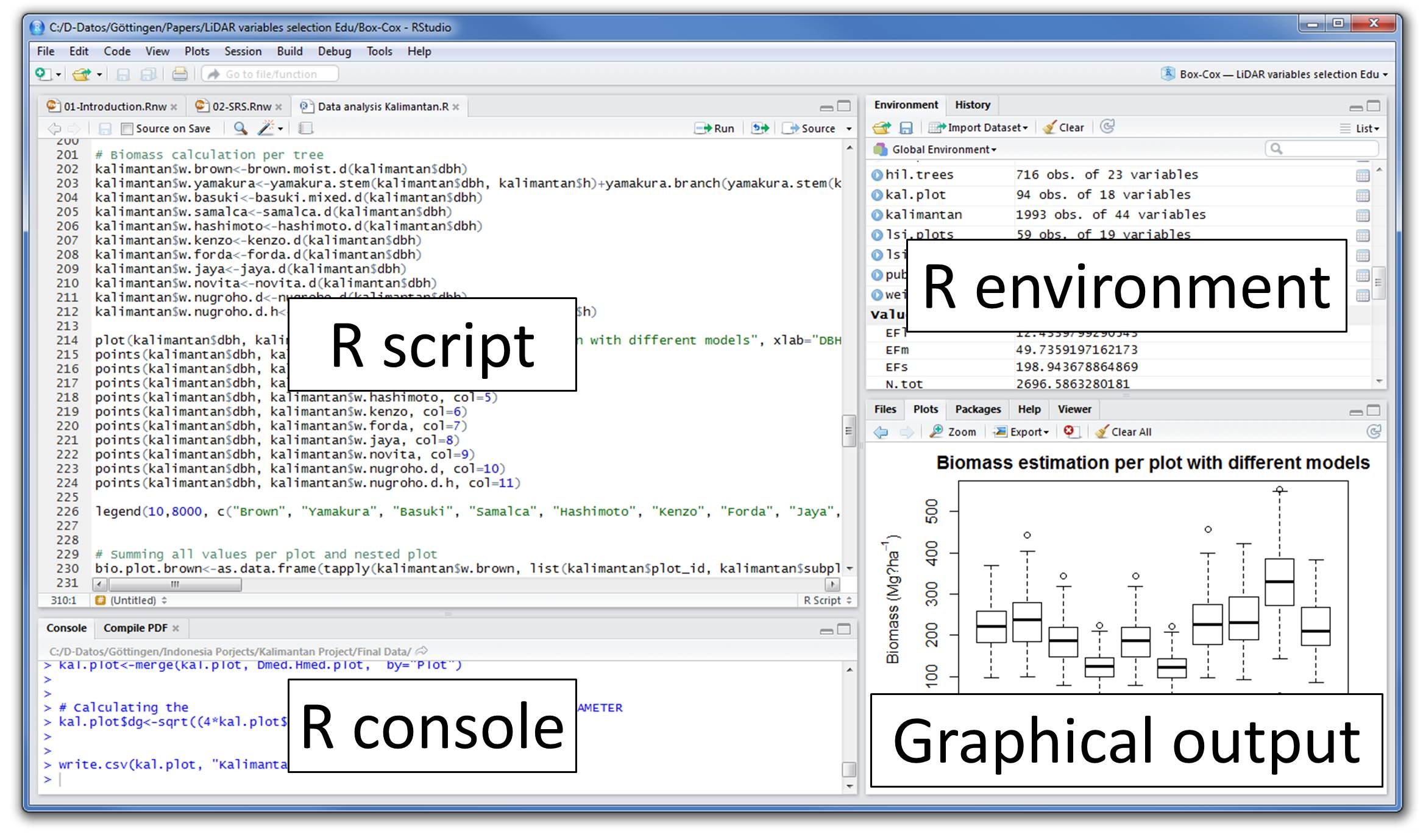
Rstudio
Integrated development environment (IDE) for R. Includes:
- A console
- Syntax-highlighting editor that supports direct code execution
- Tools for plotting, history, debugging and workspace management
Elements of the R language
- Vectors
- Lists
- Matrices
- Data Frames
- Tables
- Functions (including operators)
- Attributes
ArraysEnvironments
Data structure
The basic data structure in R is the vector. There are two basic types of vectors: atomic vectors and lists.
They have three common properties:
- Type,
typeof()(~ class/mode) - Length,
length()(number of elements) - Attributes,
attributes()(metadata)
They differ in the types of their elements: all elements of an atomic vector must be the same type, whereas the elements of a list can have different types.
| Homogeneous | Heterogeneous | |
|---|---|---|
| 1d | Atomic vector | List |
| 2d | Matrix | Data frame |
| nd | Array |
R has no 0-dimensional elements (scalars). Individual numbers or strings are actually vectors of length one.
Atomic vectors
Types of atomic vectors:
- Logical (boolean)
- Integer
- Numeric (double)
- Character
Vectors are built using c():
x <- 1
x1 <- c(1)
all.equal(x, x1)
## [1] TRUE
class(x)
## [1] "numeric"
y <- "something"
class(y)
## [1] "character"
w <- 1L
class(w)
## [1] "integer"
z <- TRUE
class(z)
## [1] "logical"
q <- factor(1)
class(q)
## [1] "factor"
Vectors can only contain entries of the same type. Different types will be coerced to the most flexible type:
v <- c(10, 11, 12, 13)
class(v)
## [1] "numeric"
typeof(v)
## [1] "double"
is.integer(v)
## [1] FALSE
y <- c("Amazona", "Ara", "Eupsittula", "Myiopsitta")
class(y)
## [1] "character"
is.integer(y)
## [1] FALSE
x <- c(1,2,3, "Myiopsitta")
x
## [1] "1" "2" "3" "Myiopsitta"
class(x)
## [1] "character"
Missing values are specified with NA, which is a logical vector of
length 1. NA will always be coerced to the correct type if used inside
c():
v <- c(10, 11, 12, 13, NA)
class(v)
## [1] "numeric"
v <- c(letters[1:3], NA)
class(v)
## [1] "character"
Lists
Can contain objects of different classes and sizes. Lists are built using list():
l <- list(ID = c("a", "b", "c", "d", "e"), size = c(1, 2, 3, 4, 5), observed = c(FALSE, TRUE, FALSE, FALSE, FALSE))
l## $ID
## [1] "a" "b" "c" "d" "e"
##
## $size
## [1] 1 2 3 4 5
##
## $observed
## [1] FALSE TRUE FALSE FALSE FALSEclass(l)## [1] "list"str(l)## List of 3
## $ ID : chr [1:5] "a" "b" "c" "d" ...
## $ size : num [1:5] 1 2 3 4 5
## $ observed: logi [1:5] FALSE TRUE FALSE FALSE FALSE… and dimensions:
l <- list(ID = c("a", "b", "c", "d", "e"), size = c(1, 2, 3, 4, 5, 6), observed = c(FALSE, TRUE, FALSE, FALSE, FALSE), l)
str(l)## List of 4
## $ ID : chr [1:5] "a" "b" "c" "d" ...
## $ size : num [1:6] 1 2 3 4 5 6
## $ observed: logi [1:5] FALSE TRUE FALSE FALSE FALSE
## $ :List of 3
## ..$ ID : chr [1:5] "a" "b" "c" "d" ...
## ..$ size : num [1:5] 1 2 3 4 5
## ..$ observed: logi [1:5] FALSE TRUE FALSE FALSE FALSEl2 <- list(l, l)
str(l2)## List of 2
## $ :List of 4
## ..$ ID : chr [1:5] "a" "b" "c" "d" ...
## ..$ size : num [1:6] 1 2 3 4 5 6
## ..$ observed: logi [1:5] FALSE TRUE FALSE FALSE FALSE
## ..$ :List of 3
## .. ..$ ID : chr [1:5] "a" "b" "c" "d" ...
## .. ..$ size : num [1:5] 1 2 3 4 5
## .. ..$ observed: logi [1:5] FALSE TRUE FALSE FALSE FALSE
## $ :List of 4
## ..$ ID : chr [1:5] "a" "b" "c" "d" ...
## ..$ size : num [1:6] 1 2 3 4 5 6
## ..$ observed: logi [1:5] FALSE TRUE FALSE FALSE FALSE
## ..$ :List of 3
## .. ..$ ID : chr [1:5] "a" "b" "c" "d" ...
## .. ..$ size : num [1:5] 1 2 3 4 5
## .. ..$ observed: logi [1:5] FALSE TRUE FALSE FALSE FALSE
Names
Vectors can be named in three ways:
- When creating it:
x <- c(a = 1, b = 2, c = 3) - By modifying an existing vector in place:
x <- 1:3;names(x) <- c("a", "b", "c")Or:x <- 1:3;names(x)[[1]] <- c("a") - By creating a modified copy of a vector:
x <- setNames(1:3, c("a", "b", "c"))
y <- c(a = 1, 2, 3)
names(y)## [1] "a" "" ""v <- c(1, 2, 3)
names(v) <- c('a')
names(v)## [1] "a" NA NAz <- setNames(1:3, c("a", "b", "c"))
names(z)## [1] "a" "b" "c"
Factors
Attributes are used to define factors. A factor is a vector that can contain only predefined values, and is used to store categorical data.
Factors are built on top of integer vectors using two attributes:
- class “factor”: makes them behave differently from regular integer vectors
- levels: defines the set of allowed values
x <- factor(c("a", "b", "b", "a"))
x## [1] a b b a
## Levels: a blevels(x)## [1] "a" "b"str(x)## Factor w/ 2 levels "a","b": 1 2 2 1
Factors look like character vectors but they are actually integers:
x <- factor(c("a", "b", "b", "a"))
c(x)## [1] a b b a
## Levels: a b
Matrices
All entries are of the same type:
m <- matrix(c(1, 2, 3, 11, 12, 13), nrow = 2)
dim(m)## [1] 2 3m## [,1] [,2] [,3]
## [1,] 1 3 12
## [2,] 2 11 13class(m)## [1] "matrix" "array"m <- matrix(c(1, 2, 3, 11, 12,"13"), nrow = 2)
m## [,1] [,2] [,3]
## [1,] "1" "3" "12"
## [2,] "2" "11" "13"
Can be created by modifying the dimension attribute:
c <- 1:6
is.matrix(c)## [1] FALSEattributes(c)## NULLdim(c) <- c(3, 2)
c## [,1] [,2]
## [1,] 1 4
## [2,] 2 5
## [3,] 3 6is.matrix(c)## [1] TRUEattributes(c)## $dim
## [1] 3 2
Data frames
Special case of lists. Can contain entries of different types:
m <- data.frame(ID = c("a", "b", "c", "d", "e"), size = c(1, 2, 3, 4, 5), observed = c(FALSE, TRUE, FALSE, FALSE, FALSE))
dim(m)## [1] 5 3m## ID size observed
## 1 a 1 FALSE
## 2 b 2 TRUE
## 3 c 3 FALSE
## 4 d 4 FALSE
## 5 e 5 FALSEclass(m)## [1] "data.frame"is.data.frame(m)## [1] TRUEis.list(m)## [1] TRUEstr(m)## 'data.frame': 5 obs. of 3 variables:
## $ ID : chr "a" "b" "c" "d" ...
## $ size : num 1 2 3 4 5
## $ observed: logi FALSE TRUE FALSE FALSE FALSE
But vectors should have the same length:
m <- data.frame(ID = c("a", "b", "c", "d", "e"), size = c(1, 2, 3, 4, 5, 6), observed = c(FALSE, TRUE, FALSE, FALSE, FALSE))## Error in data.frame(ID = c("a", "b", "c", "d", "e"), size = c(1, 2, 3, : arguments imply differing number of rows: 5, 6
Data subsetting
Indexing
Elements within objects can be called by indexing. To subset a vector simply call the object position using square brackets:
x <- c(1, 3, 4, 10, 15, 20, 50, 1, 6)
x[1]## [1] 1x[2]## [1] 3x[2:3]## [1] 3 4x[c(1,3)]## [1] 1 4
Elements can be removed in the same way:
x[-1]## [1] 3 4 10 15 20 50 1 6x[-c(1,3)]## [1] 3 10 15 20 50 1 6
Matrices and data frames required 2 indices
[row, column]:
m <- matrix(c(1, 2, 3, 11, 12, 13), nrow = 2)
m[1, ]## [1] 1 3 12m[, 1]## [1] 1 2m[1, 1]## [1] 1m[-1, ]## [1] 2 11 13m[, -1]## [,1] [,2]
## [1,] 3 12
## [2,] 11 13m[-1, -1]## [1] 11 13df <- data.frame(family = c("Psittacidae", "Trochilidae",
"Psittacidae"),
genus = c("Amazona", "Phaethornis", "Ara"),
species = c("aestiva", "philippii", "ararauna"))
df## family genus species
## 1 Psittacidae Amazona aestiva
## 2 Trochilidae Phaethornis philippii
## 3 Psittacidae Ara araraunadf[1, ]## family genus species
## 1 Psittacidae Amazona aestivadf[, 1]## [1] "Psittacidae" "Trochilidae" "Psittacidae"df[1, 1]## [1] "Psittacidae"df[-1, ]## family genus species
## 2 Trochilidae Phaethornis philippii
## 3 Psittacidae Ara araraunadf[, -1]## genus species
## 1 Amazona aestiva
## 2 Phaethornis philippii
## 3 Ara araraunadf[-1, -1]## genus species
## 2 Phaethornis philippii
## 3 Ara araraunadf[,"family"]## [1] "Psittacidae" "Trochilidae" "Psittacidae"df[,c("family", "genus")]## family genus
## 1 Psittacidae Amazona
## 2 Trochilidae Phaethornis
## 3 Psittacidae Ara
Lists require 1 index within double square brackets
[[index]]:
l <- list(ID = c("a", "b", "c", "d", "e"), size = c(1, 2, 3, 4, 5), observed = c(FALSE, TRUE, FALSE, FALSE, FALSE))
l[[1]]## [1] "a" "b" "c" "d" "e"l[[3]]## [1] FALSE TRUE FALSE FALSE FALSE
Elements within lists can also be subset in the same string of code:
l[[1]][1:2]## [1] "a" "b"l[[3]][2]## [1] TRUE
Exploring objects
str(df)## 'data.frame': 3 obs. of 3 variables:
## $ family : chr "Psittacidae" "Trochilidae" "Psittacidae"
## $ genus : chr "Amazona" "Phaethornis" "Ara"
## $ species: chr "aestiva" "philippii" "ararauna"names(df)## [1] "family" "genus" "species"dim(df)## [1] 3 3nrow(df)## [1] 3ncol(df)## [1] 3head(df)## family genus species
## 1 Psittacidae Amazona aestiva
## 2 Trochilidae Phaethornis philippii
## 3 Psittacidae Ara araraunatail(df)## family genus species
## 1 Psittacidae Amazona aestiva
## 2 Trochilidae Phaethornis philippii
## 3 Psittacidae Ara araraunatable(df$genus)##
## Amazona Ara Phaethornis
## 1 1 1typeof(df)## [1] "list"View(df)
Exercise
Using the example data
iristo create a data subset with only the observations of the species ‘setosa’Now create a data subset containing the observations of both ‘setosa’ and ‘versicolor’
Also with
iriscreate a data subset with the observations for whichiris$Sepal.lengthis higher than 6How many observations have a sepal length higher than 6?
Functions
All functions are created by the function function() and
follow the same structure:
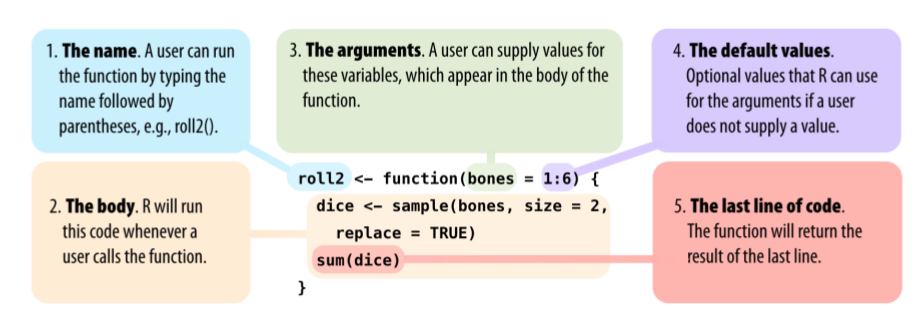
* Modified from Grolemund 2014
R comes with many functions that you can use to do sophisticated tasks:
# built in functions
bi <- builtins()
length(bi)## [1] 1370sample(bi, 10)## [1] "unlist" "mat.or.vec" "close"
## [4] "interaction" ".S3_methods_table" "vapply"
## [7] "dimnames" "digamma" "source"
## [10] ".BaseNamespaceEnv"
Operators are functions:
1 + 1## [1] 2'+'(1, 1)## [1] 22 * 3## [1] 6'*'(2, 3)## [1] 6
Most commonly used R operators
Arithmetic operators:
| Operator | Description |
|---|---|
| + | addition |
| - | subtraction |
| * | multiplication |
| / | division |
| ^ or ** | exponent |
| x %% y | modulus (x mod y) |
| x %/% y | integer division |
1 - 2## [1] -11 + 2## [1] 32 ^ 2## [1] 42 ** 2## [1] 45 %% 2## [1] 15 %/% 2## [1] 2
Logical operators:
| Operator | Description |
|---|---|
| < | less than |
| <= | less than or equal to |
| > | greater than |
| >= | greater than or equal to |
| == | exactly equal to |
| != | not equal to |
| !x | Not x |
| x | y | x OR y |
| x & y | x AND y |
| x %in% y | match |
1 < 2 ## [1] TRUE1 > 2 ## [1] FALSE1 <= 2 ## [1] TRUE1 == 2## [1] FALSE1 != 2## [1] TRUE1 > 2 ## [1] FALSE5 %in% 1:6## [1] TRUE5 %in% 1:4## [1] FALSE
Most functions are vectorized:
1:6 * 1:6
* Modified from Grolemund & Wickham 2017
## [1] 1 4 9 16 25 361:6 - 1:6## [1] 0 0 0 0 0 0R recycles vectors of unequal length:
1:6 * 1:5
* Modified from Grolemund & Wickham 2017
## Warning in 1:6 * 1:5: longitud de objeto mayor no es múltiplo de la longitud de
## uno menor## [1] 1 4 9 16 25 61:6 + 1:5## Warning in 1:6 + 1:5: longitud de objeto mayor no es múltiplo de la longitud de
## uno menor## [1] 2 4 6 8 10 7
Style matters
Based on google’s R Style Guide
File names
File names should end in .R and, of course, be meaningful:
- GOOD: predict_ad_revenue.R
- BAD: foo.R
Object names
Variables and functions:
- Lowercase
- Use an underscore (_) (HW style)
- Generally, nouns for variables and verbs for functions
- Strive for names that are concise and meaningful (not always easy)
- Avoid using names of existing functions of variables
- GOOD: day_one: day_1, mean.day(),
- BAD: dayOne, day1, firstDay_of.month, mean <- function(x) sum(x), c <- 10
Syntax
Spacing:
- Use spaces around operators and for argument within a function
- Always put a space after a comma, and never before (just like in regular English)
- Place a space before left parenthesis, except in a function call
- GOOD:
a <- rnorm(n = 10, sd = 10, mean = 1)
tab.prior <- table(df[df$days.from.opt < 0, "campaign.id"])
total <- sum(x[, 1])
total <- sum(x[1, ])
if (debug)
mean(1:10)
- BAD:
a<-rnorm(n=10,sd=10,mean=1)
tab.prior <- table(df[df$days.from.opt<0, "campaign.id"]) # Needs spaces around '<'
tab.prior <- table(df[df$days.from.opt < 0,"campaign.id"]) # Needs a space after the comma
tab.prior<- table(df[df$days.from.opt < 0, "campaign.id"]) # Needs a space before <-
tab.prior<-table(df[df$days.from.opt < 0, "campaign.id"]) # Needs spaces around <-
total <- sum(x[,1]) # Needs a space after the comma
total <- sum(x[ ,1]) # Needs a space after the comma, not before
if(debug) # Needs a space before parenthesis
mean (1:10) # ) # Extra space before parenthesis
Curly braces:
- An opening curly brace should never go on its own line
- Closing curly brace should always go on its own line
- You may omit curly braces when a block consists of a single statement
- GOOD:
if (is.null(ylim)) {
ylim <- c(0, 0.06)
}
if (is.null(ylim))
ylim <- c(0, 0.06)
- BAD:
if (is.null(ylim)) ylim <- c(0, 0.06)
if (is.null(ylim)) {ylim <- c(0, 0.06)}
if (is.null(ylim)) {
ylim <- c(0, 0.06)
}
Assigments:
- Use <-, not =
- GOOD:
x <- 5
- BAD:
x = 5
Commenting guidelines:
- Comment your code
- Entire commented lines should begin with # and one space
- Short comments can be placed after code preceded by two spaces, #, and then one space
# Create histogram of frequency of campaigns by pct budget spent.
hist(df$pct.spent,
breaks = "scott", # method for choosing number of buckets
main = "Histogram: fraction budget spent by campaignid",
xlab = "Fraction of budget spent",
ylab = "Frequency (count of campaignids)")
General Layout and Ordering (google style):
- Copyright statement comment (?)
- Author comment
- File description comment, including purpose of program, inputs, and outputs
- source() and library() statements
- Function definitions
- Executed statements, if applicable (e.g., print, plot)
R documentation
Most R resources are extremely well documented. So the first source for help you should go to when writting R code is the R documention itself. All packages are documented in the same standard way. Getting familiar with the format can simplify things a lot.
Package documentation

Reference manuals
Reference manuals are collections of the documentation for all functions in a package (only 1 per package):
Function documentation
All functions (default or from loaded packages) must have a documentation that follows a standard format:
?mean
help("mean")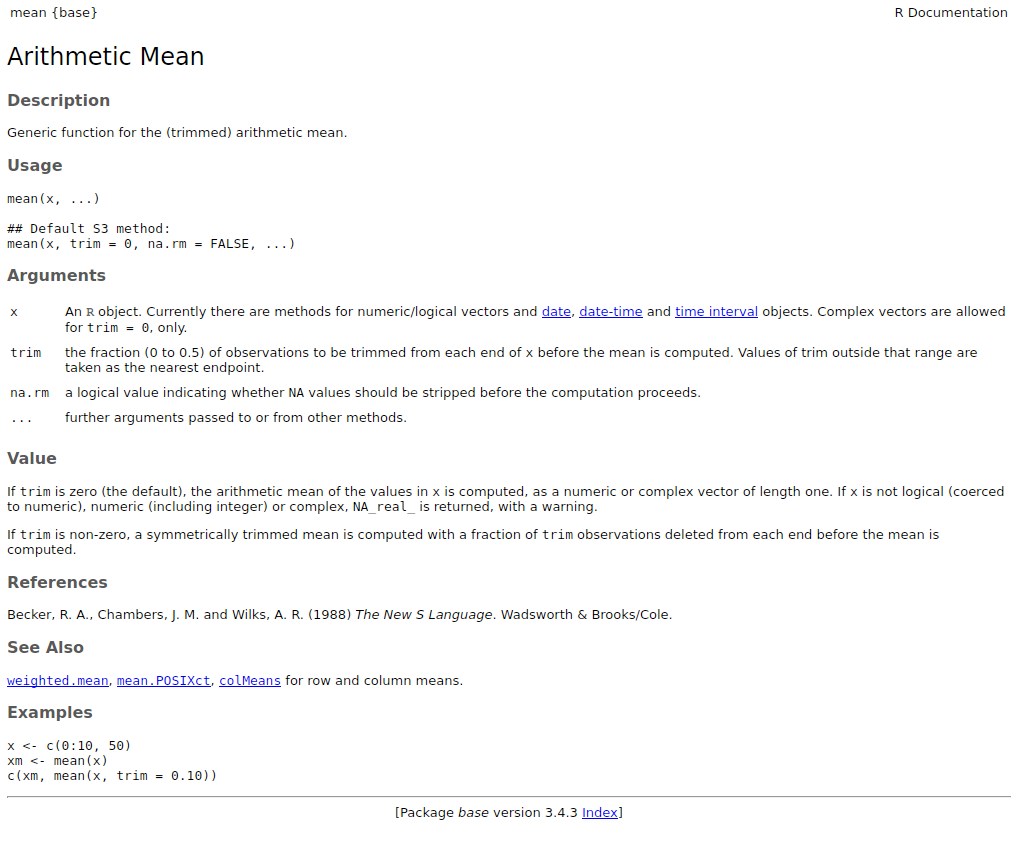
This documentation can also be shown in Rstudio by pressing
F1 when the cursor is on the function name
If you don’t recall the function name try apropos():
apropos("mean")## [1] ".colMeans" ".rowMeans" "colMeans" "kmeans"
## [5] "mean" "mean.Date" "mean.default" "mean.difftime"
## [9] "mean.POSIXct" "mean.POSIXlt" "rowMeans" "weighted.mean"
Vignettes
Vignettes are illustrative documents or study cases detailing the use of a package (optional, can be several per package).
Vignettes can be called directly from R:
vgn <- browseVignettes() vignette()They should also be listed in the package CRAN page.
Demonstrations
Packages may also include extended code demonstrations (‘demos’). To
list demos in a package run demo("package name"):
demo(package="stats")
# call demo directly
demo("nlm")
Exercise
What does the function
cut()do?What is the
breaksargument incut()used for?Run the first 4 lines of code in the examples supplied in the
cut()documentationHow many vignettes does the package warbleR has?
References
- Advanced R, H Wickham
- Google’s R
Style Guide
- Hands-On Programming with R (Grolemund, 2014)
Session information
## R version 4.1.1 (2021-08-10)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 20.04.2 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
##
## locale:
## [1] LC_CTYPE=es_ES.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=es_CR.UTF-8 LC_COLLATE=es_ES.UTF-8
## [5] LC_MONETARY=es_CR.UTF-8 LC_MESSAGES=es_ES.UTF-8
## [7] LC_PAPER=es_CR.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=es_CR.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] kableExtra_1.3.4 knitr_1.37
##
## loaded via a namespace (and not attached):
## [1] rstudioapi_0.13 xml2_1.3.2 magrittr_2.0.2 rvest_1.0.1
## [5] munsell_0.5.0 viridisLite_0.4.0 colorspace_2.0-3 R6_2.5.1
## [9] rlang_1.0.2 fastmap_1.1.0 stringr_1.4.0 httr_1.4.2
## [13] tools_4.1.1 webshot_0.5.2 xfun_0.30 cli_3.2.0
## [17] jquerylib_0.1.4 systemfonts_1.0.2 htmltools_0.5.2 yaml_2.3.5
## [21] digest_0.6.29 lifecycle_1.0.1 sass_0.4.0 glue_1.6.2
## [25] evaluate_0.15 rmarkdown_2.10 stringi_1.7.6 compiler_4.1.1
## [29] bslib_0.2.5.1 scales_1.1.1 svglite_2.0.0 jsonlite_1.7.2